Single-cell omics enables the analysis of gene expression, protein levels, and metabolic activity at the individual cell level, uncovering cellular heterogeneity within complex tissues that bulk omics often masks. Bulk omics aggregates data from millions of cells, providing average molecular profiles that can obscure rare cell populations and subtle regulatory mechanisms. Advances in single-cell technologies offer unprecedented resolution for understanding cellular diversity, disease progression, and therapeutic targets in biological engineering.
Table of Comparison
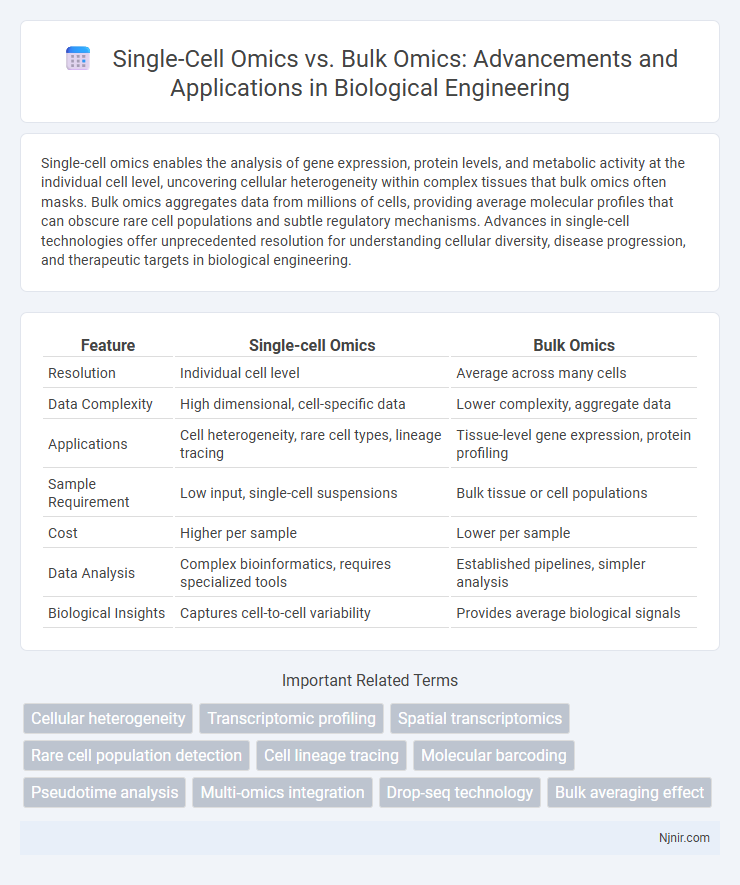
| Feature | Single-cell Omics | Bulk Omics |
|---|---|---|
| Resolution | Individual cell level | Average across many cells |
| Data Complexity | High dimensional, cell-specific data | Lower complexity, aggregate data |
| Applications | Cell heterogeneity, rare cell types, lineage tracing | Tissue-level gene expression, protein profiling |
| Sample Requirement | Low input, single-cell suspensions | Bulk tissue or cell populations |
| Cost | Higher per sample | Lower per sample |
| Data Analysis | Complex bioinformatics, requires specialized tools | Established pipelines, simpler analysis |
| Biological Insights | Captures cell-to-cell variability | Provides average biological signals |
Introduction to Omics Technologies in Biological Engineering
Single-cell omics enables the analysis of genomes, transcriptomes, and proteomes at the individual cell level, revealing cellular heterogeneity critical for precise biological engineering applications. Bulk omics aggregates molecular data from millions of cells, providing average profiles that may overlook rare cell populations and unique cellular states. Integrating single-cell and bulk omics technologies enhances the resolution and accuracy of biological insights, facilitating advanced biomarker discovery and targeted therapeutic development.
Fundamentals of Single-cell Omics
Single-cell omics enables analysis of genomic, transcriptomic, proteomic, or epigenomic data at the resolution of individual cells, revealing cellular heterogeneity that bulk omics, which averages signals across many cells, cannot capture. Techniques such as single-cell RNA sequencing (scRNA-seq) provide insights into cell-specific gene expression profiles, facilitating the study of complex tissues, developmental processes, and disease microenvironments. This fundamental approach allows for identification of rare cell populations and dynamic cellular states critical for precision medicine and biological discovery.
Overview of Bulk Omics Approaches
Bulk omics approaches analyze aggregated molecular data from mixed cell populations, providing comprehensive insights into gene expression, protein abundance, and metabolic profiles at the tissue or organism level. Techniques such as bulk RNA sequencing, proteomics, and metabolomics enable identification of global biological trends and pathways but lack resolution of cell-to-cell variability. This limits their ability to detect rare cell types or subtle cellular heterogeneity critical for understanding complex diseases and cellular functions.
Resolution Differences: Single-cell vs Bulk Omics
Single-cell omics provides high-resolution insights by analyzing gene expression, epigenetic modifications, or protein profiles at the individual cell level, revealing cellular heterogeneity within complex tissues. Bulk omics aggregates molecular data from large populations of cells, offering an average signal that can mask distinct cellular subpopulations and rare cell types. This difference in resolution makes single-cell omics critical for studying developmental processes, disease mechanisms, and tumor microenvironments where cellular diversity drives biological function.
Applications in Disease Diagnostics
Single-cell omics enables precise identification of cellular heterogeneity within tissues, providing critical insights for early cancer detection and personalized treatment strategies. Bulk omics aggregates data from numerous cells, which can obscure rare but clinically significant cell populations important in diseases like neurodegeneration and infectious diseases. Advanced single-cell transcriptomics and proteomics are transforming diagnostics by revealing disease mechanisms at the individual cell level, facilitating targeted therapies and improved prognostic accuracy.
Insights into Cellular Heterogeneity
Single-cell omics provides unparalleled resolution to analyze cellular heterogeneity by profiling gene expression, epigenetics, and proteomics at the individual cell level, revealing diverse cell populations and rare cell types often masked in bulk omics. Bulk omics averages signals across thousands or millions of cells, which can obscure critical variations in cellular states and functions. Leveraging single-cell omics enhances understanding of tissue complexity, disease mechanisms, and therapeutic responses by capturing distinct cellular behaviors within heterogeneous samples.
Data Integration and Analytical Challenges
Single-cell omics offers high-resolution data capturing cellular heterogeneity, whereas bulk omics aggregates signals across diverse cell populations, masking individual cell variations. Integrating single-cell and bulk omics data requires advanced computational frameworks to align disparate data modalities, scales, and noise levels, often leveraging machine learning algorithms and statistical models for effective data fusion. Analytical challenges include managing batch effects, addressing sparsity in single-cell datasets, and developing scalable pipelines capable of interpreting complex multi-omics interactions across heterogeneous biological contexts.
Technological Advances Driving Omics Research
Single-cell omics technologies enable high-resolution analysis of individual cells, overcoming the averaging limitations of bulk omics by capturing cellular heterogeneity and rare cell populations. Innovations in microfluidics, next-generation sequencing, and bioinformatics algorithms have dramatically increased throughput and accuracy in single-cell RNA sequencing and proteomics. These advancements allow researchers to dissect complex biological systems with unprecedented detail, enhancing insights into development, disease progression, and therapeutic responses.
Comparative Advantages and Limitations
Single-cell omics enables high-resolution analysis of cellular heterogeneity by profiling individual cells, revealing rare cell types and dynamic states that bulk omics averages out. Bulk omics provides robust, cost-effective insights into overall tissue or sample-level molecular profiles but lacks spatial and cell-specific data crucial for understanding complex biological systems. Single-cell techniques face challenges like higher noise, technical variability, and increased computational demands, whereas bulk methods benefit from greater data stability and simpler analysis pipelines.
Future Perspectives in Omics-driven Biological Engineering
Single-cell omics enable precise profiling of cellular heterogeneity, revealing rare cell populations and dynamic biological processes that bulk omics average out. Advances in multi-omics integration and high-throughput single-cell techniques will drive innovations in synthetic biology, regenerative medicine, and personalized therapeutics. Future omics-driven biological engineering will leverage single-cell resolution to design more accurate gene circuits, optimize metabolic pathways, and enhance biomolecular engineering with unprecedented specificity and efficiency.
Cellular heterogeneity
Single-cell omics captures cellular heterogeneity by profiling individual cells' molecular signatures, whereas bulk omics averages signals across mixed cell populations, masking distinct cellular differences.
Transcriptomic profiling
Single-cell transcriptomic profiling reveals cellular heterogeneity and rare cell populations by analyzing gene expression at individual cell resolution, whereas bulk transcriptomics averages gene expression across mixed cell populations, masking cellular diversity.
Spatial transcriptomics
Spatial transcriptomics in single-cell omics enables precise gene expression mapping at cellular resolution, outperforming bulk omics by preserving spatial context and revealing cell-type-specific interactions.
Rare cell population detection
Single-cell omics enables precise detection and characterization of rare cell populations by analyzing individual cellular heterogeneity, while bulk omics averages signals across millions of cells, often masking rare cell-specific information.
Cell lineage tracing
Single-cell omics enables precise cell lineage tracing by analyzing individual cell genomes, transcriptomes, or epigenomes, unlike bulk omics that average signals across heterogeneous cell populations.
Molecular barcoding
Molecular barcoding in single-cell omics enables precise identification and quantification of individual cell transcripts, offering higher resolution than bulk omics which averages signals across heterogeneous cell populations.
Pseudotime analysis
Single-cell omics enables precise Pseudotime analysis by capturing cellular heterogeneity and dynamic gene expression trajectories, unlike Bulk omics which averages signals across mixed cell populations, obscuring temporal progression.
Multi-omics integration
Multi-omics integration in single-cell omics enables high-resolution insights into cellular heterogeneity by combining transcriptomic, proteomic, and epigenomic data, surpassing bulk omics which averages signals across mixed cell populations.
Drop-seq technology
Drop-seq technology enables high-throughput single-cell omics by encapsulating individual cells with uniquely barcoded beads, allowing precise transcriptomic profiling that overcomes the averaging limitations of bulk omics.
Bulk averaging effect
Bulk omics datasets average molecular signals across diverse cell populations, obscuring cellular heterogeneity that single-cell omics can reveal.
Single-cell omics vs Bulk omics Infographic
 njnir.com
njnir.com