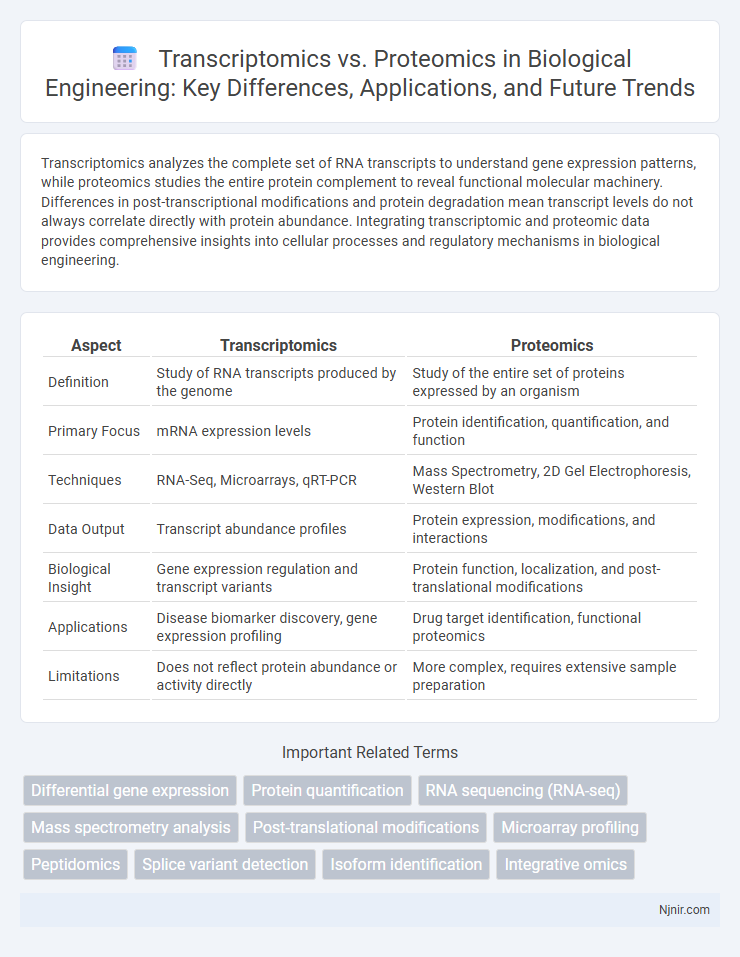
Transcriptomics analyzes the complete set of RNA transcripts to understand gene expression patterns, while proteomics studies the entire protein complement to reveal functional molecular machinery. Differences in post-transcriptional modifications and protein degradation mean transcript levels do not always correlate directly with protein abundance. Integrating transcriptomic and proteomic data provides comprehensive insights into cellular processes and regulatory mechanisms in biological engineering.
Table of Comparison
| Aspect | Transcriptomics | Proteomics |
|---|---|---|
| Definition | Study of RNA transcripts produced by the genome | Study of the entire set of proteins expressed by an organism |
| Primary Focus | mRNA expression levels | Protein identification, quantification, and function |
| Techniques | RNA-Seq, Microarrays, qRT-PCR | Mass Spectrometry, 2D Gel Electrophoresis, Western Blot |
| Data Output | Transcript abundance profiles | Protein expression, modifications, and interactions |
| Biological Insight | Gene expression regulation and transcript variants | Protein function, localization, and post-translational modifications |
| Applications | Disease biomarker discovery, gene expression profiling | Drug target identification, functional proteomics |
| Limitations | Does not reflect protein abundance or activity directly | More complex, requires extensive sample preparation |
Introduction to Transcriptomics and Proteomics
Transcriptomics involves the comprehensive analysis of RNA transcripts produced by the genome under specific conditions, providing insights into gene expression and regulation. Proteomics focuses on the large-scale study of proteins, their structures, functions, and interactions, revealing the functional output of gene expression. Both fields use advanced technologies like RNA sequencing and mass spectrometry, enabling a deeper understanding of cellular mechanisms and molecular biology.
Core Concepts: mRNA vs Protein Expression
Transcriptomics analyzes mRNA expression levels to reveal gene activity, reflecting which genes are transcribed under specific conditions. Proteomics studies protein expression, modification, and interaction, providing insights into functional biomolecules directly responsible for cellular processes. While transcriptomics measures potential protein synthesis, proteomics captures actual protein abundance and activity, highlighting post-transcriptional regulation effects.
Technological Platforms: Sequencing vs Mass Spectrometry
Transcriptomics primarily utilizes high-throughput sequencing technologies such as RNA-Seq to quantify and analyze RNA transcripts, providing insights into gene expression profiles and alternative splicing events. Proteomics relies on mass spectrometry platforms, including tandem MS/MS and MALDI-TOF, to identify and quantify proteins, post-translational modifications, and protein-protein interactions. The integration of sequencing and mass spectrometry technologies enhances comprehensive molecular understanding by linking gene expression with functional protein products.
Data Output and Interpretation in Transcriptomics and Proteomics
Transcriptomics generates large datasets of RNA sequences reflecting gene expression levels across different conditions and time points, requiring bioinformatics tools for quantification and differential expression analysis. Proteomics produces complex data on protein identities, abundances, modifications, and interactions through mass spectrometry or antibody-based methods, presenting challenges in peptide mapping and functional annotation. Interpretation in transcriptomics centers on mRNA abundance as a proxy for gene activity, while proteomics provides direct insights into functional proteins and post-translational modifications, crucial for understanding cellular mechanisms.
Resolution and Sensitivity: Comparing Analytical Depth
Transcriptomics offers high-resolution analysis of gene expression by quantifying mRNA levels with sensitivity to detect low-abundance transcripts, enabling detailed insights into cellular functions. Proteomics provides comprehensive profiling of protein abundance, modifications, and interactions but faces challenges in sensitivity due to protein complexity and dynamic range, often requiring advanced mass spectrometry techniques for deep analytical coverage. The analytical depth in transcriptomics surpasses proteomics in quantifying gene expression precisely, while proteomics excels in functional characterization of the proteome despite comparatively lower sensitivity and resolution.
Applications in Biological Engineering
Transcriptomics analyzes RNA transcripts to reveal gene expression patterns, enabling precise control of cellular functions in biological engineering. Proteomics studies the entire protein set, providing insights into protein interactions and post-translational modifications critical for designing synthetic biological systems. Both approaches optimize metabolic engineering, biomarker discovery, and therapeutic development by integrating molecular-level data for enhanced system performance and stability.
Advantages and Limitations of Each Approach
Transcriptomics offers a comprehensive analysis of gene expression by quantifying mRNA levels, enabling the identification of active genes and regulatory mechanisms with high sensitivity, but it does not provide direct information about protein abundance or post-translational modifications. Proteomics directly measures protein expression, structure, and modifications, providing insights into functional biomolecules and cellular processes that transcriptomics cannot capture; however, it faces challenges such as lower sensitivity, dynamic range limitations, and the complexity of protein extraction and identification. Integrating transcriptomics and proteomics enhances understanding of biological systems by compensating for the limitations of each approach and offering a multi-layered perspective on molecular function and regulation.
Integrative Multi-Omics Analysis
Integrative multi-omics analysis combines transcriptomics and proteomics to provide a comprehensive understanding of cellular functions by correlating gene expression data with protein abundance and modifications. Transcriptomics captures mRNA levels reflecting gene activity, while proteomics identifies the functional protein output, revealing post-transcriptional regulation and protein-protein interactions. This holistic approach enhances biomarker discovery and pathway analysis by integrating complementary data layers for systems biology insights.
Future Trends in Systems Biology
Transcriptomics and proteomics are pivotal in unraveling biological complexity, with future trends emphasizing integrative multi-omics approaches to enhance systems biology insights. Advances in single-cell RNA sequencing and mass spectrometry are driving high-resolution temporal and spatial mapping of gene and protein expression. Emerging computational frameworks leveraging artificial intelligence enable predictive modeling of dynamic cellular networks, accelerating discoveries in precision medicine and synthetic biology.
Choosing the Right Approach for Your Research
Choosing between transcriptomics and proteomics depends on whether the focus is on gene expression or functional protein activity. Transcriptomics analyzes mRNA levels to understand gene regulation, while proteomics examines protein abundance, modifications, and interactions that directly impact cellular processes. Researchers must consider study objectives, sample type, and available technology to select the most informative method for their specific biological question.
Differential gene expression
Differential gene expression analysis in transcriptomics quantifies mRNA levels to reveal gene activity changes, while proteomics measures corresponding protein abundance and modifications, providing complementary insights into cellular function and regulation.
Protein quantification
Protein quantification in proteomics provides direct measurement of protein abundance and modifications, offering more accurate insights into cellular functions compared to transcriptomics, which analyzes mRNA levels as indirect indicators.
RNA sequencing (RNA-seq)
RNA sequencing (RNA-seq) provides high-resolution transcriptomic data by quantifying gene expression levels, enabling detailed analysis of RNA molecules that complements proteomics in understanding cellular function and regulation.
Mass spectrometry analysis
Mass spectrometry analysis in transcriptomics quantifies RNA molecules to study gene expression, whereas in proteomics it identifies and measures protein abundance and modifications to elucidate functional cellular processes.
Post-translational modifications
Post-translational modifications in proteomics provide critical functional insights beyond transcriptomics by directly revealing protein activity, localization, and interaction dynamics that mRNA expression levels alone cannot capture.
Microarray profiling
Microarray profiling in transcriptomics enables high-throughput analysis of gene expression levels by measuring mRNA abundance, whereas proteomics focuses on protein identification and quantification through techniques like mass spectrometry, providing complementary insights into cellular function.
Peptidomics
Peptidomics, a specialized branch of proteomics, focuses on the comprehensive analysis of endogenous peptides to reveal functional biomolecules and post-translational modifications beyond the scope of traditional transcriptomics.
Splice variant detection
Transcriptomics enables comprehensive splice variant detection by analyzing RNA sequences, whereas proteomics identifies resulting protein isoforms but offers limited sensitivity for directly detecting splice variants.
Isoform identification
Transcriptomics provides comprehensive isoform identification by analyzing RNA transcripts, while proteomics directly detects protein isoforms but with lower sensitivity and coverage.
Integrative omics
Integrative omics combines transcriptomics and proteomics to provide a comprehensive understanding of gene expression regulation and protein function by correlating mRNA levels with protein abundance and modifications.
Transcriptomics vs Proteomics Infographic
 njnir.com
njnir.com