Wet labs enable biological engineers to perform hands-on experiments with living organisms, chemicals, and biological samples, facilitating direct observation and manipulation of biological processes. Dry labs utilize computational tools, data analysis, and simulations to model biological systems, offering efficient hypothesis testing and prediction without physical experiments. Combining wet and dry lab approaches enhances innovation and accuracy in biological engineering research and development.
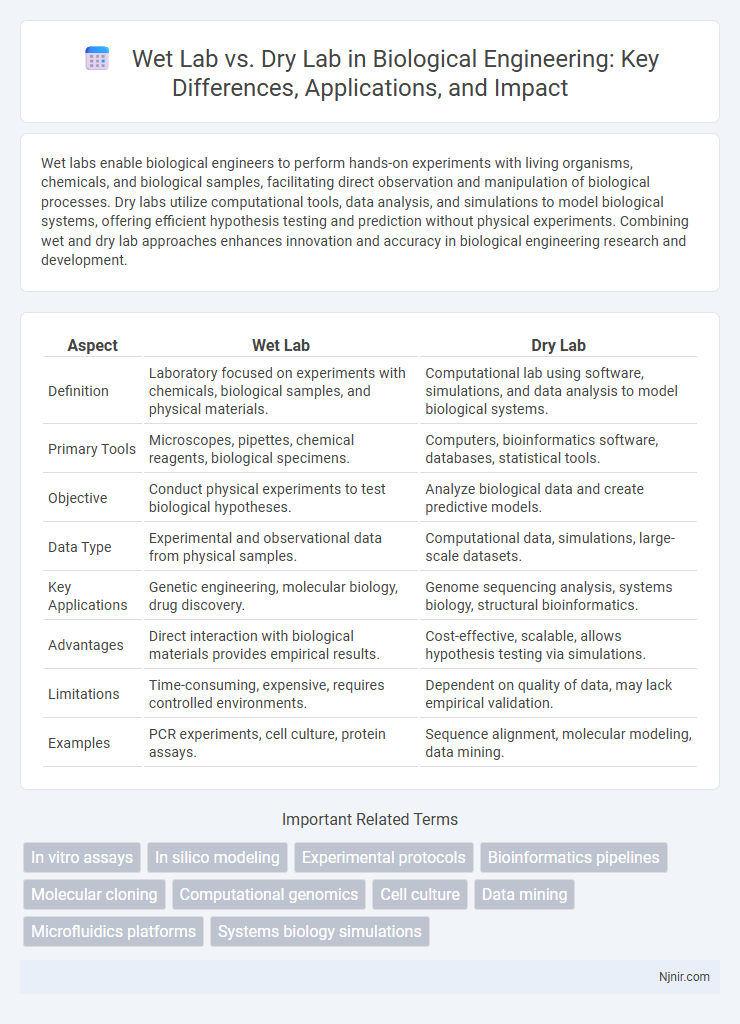
Table of Comparison
| Aspect | Wet Lab | Dry Lab |
|---|---|---|
| Definition | Laboratory focused on experiments with chemicals, biological samples, and physical materials. | Computational lab using software, simulations, and data analysis to model biological systems. |
| Primary Tools | Microscopes, pipettes, chemical reagents, biological specimens. | Computers, bioinformatics software, databases, statistical tools. |
| Objective | Conduct physical experiments to test biological hypotheses. | Analyze biological data and create predictive models. |
| Data Type | Experimental and observational data from physical samples. | Computational data, simulations, large-scale datasets. |
| Key Applications | Genetic engineering, molecular biology, drug discovery. | Genome sequencing analysis, systems biology, structural bioinformatics. |
| Advantages | Direct interaction with biological materials provides empirical results. | Cost-effective, scalable, allows hypothesis testing via simulations. |
| Limitations | Time-consuming, expensive, requires controlled environments. | Dependent on quality of data, may lack empirical validation. |
| Examples | PCR experiments, cell culture, protein assays. | Sequence alignment, molecular modeling, data mining. |
Introduction to Wet Lab and Dry Lab in Biological Engineering
Wet labs in biological engineering involve hands-on experiments using liquids, biological materials, and chemicals to study living organisms and biochemical processes. Dry labs focus on computational modeling, data analysis, and simulations to interpret biological data and predict system behaviors without physical experimentation. Combining wet and dry lab techniques enhances research efficiency and innovation in areas like genetic engineering, drug development, and systems biology.
Core Functions of Wet Labs
Wet labs specialize in conducting experiments that require direct handling of liquids, chemicals, and biological materials, enabling processes such as chemical synthesis, cell culture, and molecular biology assays. These labs are equipped with essential tools like fume hoods, pipettes, and centrifuges to support precise manipulation of substances in aqueous environments. Core functions include biochemical reactions, microbiological culturing, and sample preparation for downstream analysis, which are critical for research in biotechnology, pharmacology, and medical diagnostics.
Core Functions of Dry Labs
Dry labs primarily focus on computational analysis, data modeling, and simulation of biological processes, enabling researchers to predict outcomes without physical experiments. These labs leverage bioinformatics tools, statistical software, and machine learning algorithms to interpret complex datasets from genomics, proteomics, and metabolomics studies. This approach enhances efficiency in hypothesis testing, accelerates data-driven decision-making, and reduces the need for costly, time-intensive wet lab experiments.
Key Equipment and Tools Used
Wet labs primarily utilize equipment such as microscopes, centrifuges, pipettes, and autoclaves to perform biological, chemical, or biochemical experiments involving liquids and chemicals. Dry labs rely on computational tools, including high-performance computers, software for data analysis, modeling, and simulations, as well as databases and programming languages for processing experimental data. Both lab types emphasize specific equipment tailored to their experimental methods to ensure precise data collection and analysis.
Common Research Techniques and Workflows
Wet labs involve hands-on experiments with biological samples, chemicals, and physical materials using techniques such as pipetting, cell culturing, and chromatography, essential for molecular biology, biochemistry, and pharmacology research. Dry labs focus on computational methods, data analysis, and simulations, employing bioinformatics tools, statistical modeling, and data visualization to interpret large datasets and predict biological outcomes. Integration of wet lab experiments and dry lab analyses accelerates research workflows, enhancing hypothesis testing, data validation, and experimental design optimization.
Advantages of Wet Lab Approaches
Wet lab approaches enable direct experimentation with biological samples, allowing precise manipulation and observation of biochemical processes in real time. These methods provide tangible data on molecular interactions, enzyme activities, and cellular functions that computational models may not fully replicate. Access to authentic biological environments ensures accurate validation of hypotheses and development of practical applications in pharmacology and genetics.
Advantages of Dry Lab Approaches
Dry lab approaches offer significant advantages in terms of cost efficiency and speed, eliminating the need for physical reagents, laboratory space, and extensive safety protocols. Computational modeling and simulations enable high-throughput analysis and hypothesis testing, accelerating research in genomics, bioinformatics, and systems biology. These methods facilitate reproducibility and data sharing, enhancing collaboration and enabling large-scale data integration across diverse scientific disciplines.
Challenges and Limitations of Wet Labs
Wet labs face significant challenges such as the high costs of reagents, maintenance of delicate equipment, and strict safety protocols to handle hazardous materials. Experimental variability due to environmental factors like temperature and contamination risks can limit reproducibility and precision. Time-consuming procedures and the need for skilled personnel further constrain the scalability and efficiency of wet lab research.
Challenges and Limitations of Dry Labs
Dry labs face challenges in replicating complex biological environments accurately due to limitations in modeling dynamic biochemical interactions and cellular behaviors. Computational simulations and data analysis depend heavily on the quality and completeness of input data, which can lead to potential inaccuracies or oversights. Without physical experiments, dry labs often struggle to validate hypotheses experimentally, limiting their ability to confirm real-world biological phenomena.
Integrative Approaches: Bridging Wet Lab and Dry Lab
Integrative approaches in scientific research combine wet lab experiments, involving physical biological or chemical processes, with dry lab techniques, which utilize computational simulations and data analysis to enhance understanding. This synergy enables researchers to validate computational models experimentally and refine hypotheses through iterative feedback between hands-on experimentation and in silico predictions. Bridging wet lab and dry lab methodologies accelerates discoveries in genomics, drug development, and systems biology by leveraging complementary strengths of empirical data and computational insights.
In vitro assays
In vitro assays conducted in wet labs allow direct manipulation of biological samples with chemicals and reagents, whereas dry labs use computational models and simulations to analyze experimental data without physical experimentation.
In silico modeling
In silico modeling in dry labs enables precise simulation of biological processes using computational methods, reducing the need for time-consuming and costly experiments typically conducted in wet labs.
Experimental protocols
Wet labs involve hands-on experimental protocols using biological samples and chemical reagents, while dry labs focus on computational simulations and data analysis protocols without physical specimen handling.
Bioinformatics pipelines
Bioinformatics pipelines in dry labs process computational biological data, while wet labs generate experimental data through hands-on biological experiments essential for pipeline validation.
Molecular cloning
Molecular cloning in wet labs involves hands-on experimental procedures such as DNA extraction and ligation, while dry labs utilize computational tools for sequence analysis and primer design.
Computational genomics
Computational genomics primarily relies on dry lab techniques involving bioinformatics and data analysis, whereas wet labs focus on experimental procedures like DNA extraction and sequencing.
Cell culture
Wet labs enable hands-on cell culture experiments involving living cells and biological materials, while dry labs use computational models and simulations to analyze cell behavior without physical samples.
Data mining
Data mining in wet labs involves experimental data collection from biological samples, whereas in dry labs it focuses on computational analysis and modeling of existing datasets.
Microfluidics platforms
Microfluidics platforms in wet labs utilize physical fluids for experimental manipulation, whereas dry labs employ computer simulations and modeling to analyze microfluidic system behaviors without physical experiments.
Systems biology simulations
Systems biology simulations in dry labs leverage computational models to analyze complex biological systems, while wet labs provide empirical data through experimental procedures essential for validating these simulations.
Wet lab vs Dry lab Infographic
 njnir.com
njnir.com