Cell-free protein synthesis enables rapid and controlled production of proteins without the complexities of living cells, allowing precise manipulation of the biochemical environment for optimized yield. In vivo protein synthesis occurs within living cells, providing a natural context for post-translational modifications and protein folding but often facing limitations from cellular toxicity and resource competition. Advances in biological engineering are enhancing cell-free systems to achieve efficiencies comparable to traditional in vivo methods while offering scalability and flexibility for synthetic biology applications.
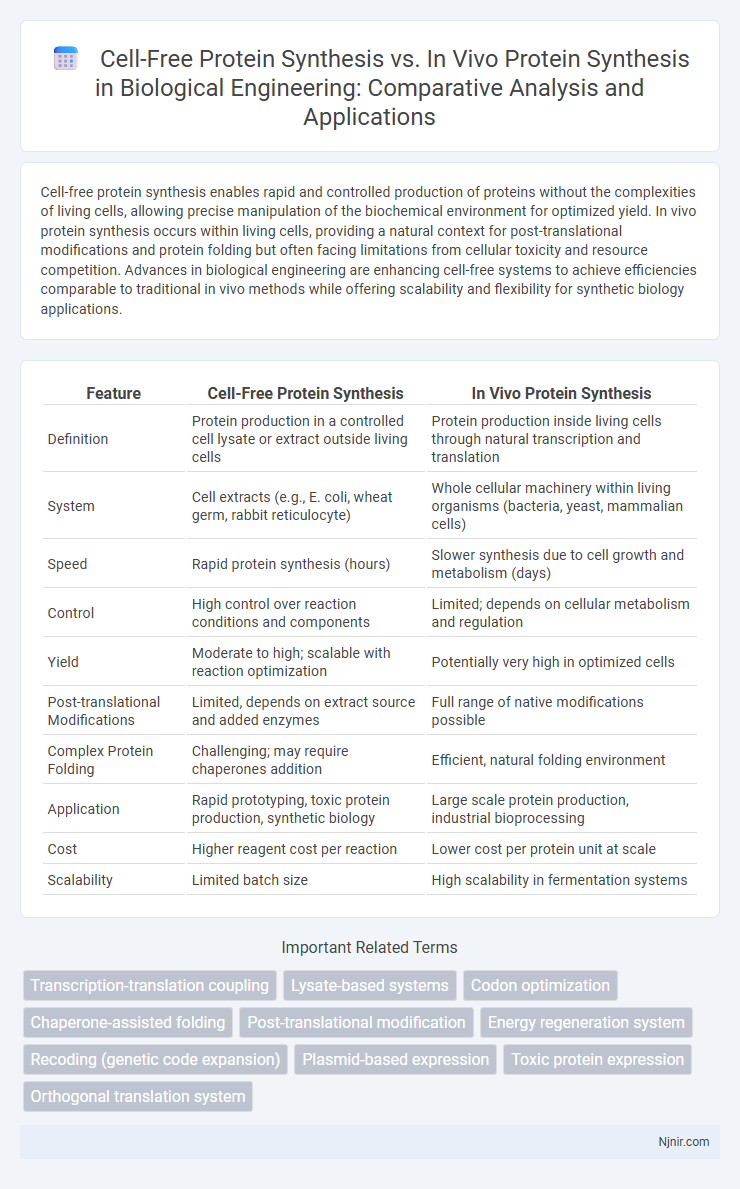
Table of Comparison
| Feature | Cell-Free Protein Synthesis | In Vivo Protein Synthesis |
|---|---|---|
| Definition | Protein production in a controlled cell lysate or extract outside living cells | Protein production inside living cells through natural transcription and translation |
| System | Cell extracts (e.g., E. coli, wheat germ, rabbit reticulocyte) | Whole cellular machinery within living organisms (bacteria, yeast, mammalian cells) |
| Speed | Rapid protein synthesis (hours) | Slower synthesis due to cell growth and metabolism (days) |
| Control | High control over reaction conditions and components | Limited; depends on cellular metabolism and regulation |
| Yield | Moderate to high; scalable with reaction optimization | Potentially very high in optimized cells |
| Post-translational Modifications | Limited, depends on extract source and added enzymes | Full range of native modifications possible |
| Complex Protein Folding | Challenging; may require chaperones addition | Efficient, natural folding environment |
| Application | Rapid prototyping, toxic protein production, synthetic biology | Large scale protein production, industrial bioprocessing |
| Cost | Higher reagent cost per reaction | Lower cost per protein unit at scale |
| Scalability | Limited batch size | High scalability in fermentation systems |
Overview of Protein Synthesis Methods
Cell-free protein synthesis enables rapid production of proteins outside living cells using cellular extracts, offering precise control over reaction conditions and incorporation of non-natural amino acids. In vivo protein synthesis occurs within living organisms, leveraging cellular machinery in a natural environment, which supports protein folding and post-translational modifications. Cell-free systems are advantageous for high-throughput screening and synthetic biology applications, while in vivo methods remain essential for producing complex proteins with proper functionality.
Principles of Cell-Free Protein Synthesis
Cell-free protein synthesis operates by harnessing the transcriptional and translational machinery extracted from cells, such as ribosomes, tRNAs, amino acids, and enzymes, in a controlled, cell-free environment. This approach allows direct manipulation of reaction conditions and substrates without the constraints of cellular membranes or metabolic regulation inherent in in vivo protein synthesis. The open system of cell-free protein synthesis enables rapid protein production, incorporation of non-natural amino acids, and high-throughput screening for synthetic biology and protein engineering applications.
Mechanisms of In Vivo Protein Synthesis
In vivo protein synthesis occurs within living cells, involving transcription of DNA into mRNA in the nucleus followed by translation of mRNA into polypeptides at ribosomes in the cytoplasm. Key molecular players include RNA polymerase for transcription and ribosomal subunits, tRNAs, and various translation factors for decoding mRNA into amino acid chains. This tightly regulated process depends on cellular machinery, energy sources like ATP and GTP, and post-translational modifications to ensure proper protein folding and function.
Comparative Efficiency: Cell-Free vs In Vivo
Cell-free protein synthesis offers rapid protein production by bypassing cellular growth and metabolism, enabling precise control over reaction conditions and high-throughput screening, which contrasts with in vivo systems that depend on cellular resources and slower expression timelines. Although in vivo protein synthesis benefits from natural protein folding machinery and post-translational modifications, cell-free systems often achieve higher yield per unit time due to optimized reaction environments and reduced degradation. The scalability and adaptability of cell-free platforms provide efficiency advantages in synthetic biology applications, despite in vivo synthesis remaining dominant for complex protein production requiring cellular contexts.
Scalability and Throughput in Protein Production
Cell-free protein synthesis offers rapid scalability with the ability to customize reaction conditions for high-throughput screening, enabling efficient protein production without the complexity of living cells. In vivo protein synthesis relies on cellular machinery, which can limit scalability due to factors such as cell growth constraints and metabolic burden, but it supports continuous protein production over extended times. High-throughput applications benefit from cell-free systems because they reduce variability and allow direct manipulation of the environment, boosting screening speed and protein yield consistency.
Applications in Synthetic Biology and Biotechnology
Cell-free protein synthesis enables rapid prototyping of genetic circuits and production of toxic or non-natural proteins difficult to express in vivo, accelerating synthetic biology applications. In vivo protein synthesis remains essential for large-scale, cost-effective production of complex proteins requiring post-translational modifications and cellular context. Both platforms complement each other, with cell-free systems offering flexibility and speed, while in vivo synthesis provides scalability and biological functionality critical for biotechnology and therapeutic development.
Flexibility in Protein Engineering and Modification
Cell-free protein synthesis offers unparalleled flexibility in protein engineering by enabling direct manipulation of reaction conditions and incorporation of non-standard amino acids without cellular toxicity constraints. In vivo protein synthesis is limited by cellular metabolic pathways and regulatory mechanisms, restricting modifications to naturally occurring amino acids and post-translational processes. Cell-free systems also allow rapid prototyping and high-throughput screening of protein variants, accelerating synthetic biology applications and therapeutic protein design.
Challenges and Limitations of Each Approach
Cell-free protein synthesis faces challenges such as limited scalability, high costs of reagents, and difficulties in replicating post-translational modifications found in natural cellular environments. In vivo protein synthesis is constrained by factors including cellular toxicity of expressed proteins, metabolic burden on host cells, and slower reaction times due to complex regulatory mechanisms. Both methods exhibit limitations in yield optimization, with cell-free systems excelling in speed and control but lagging in cost-effectiveness, while in vivo systems provide more natural folding and modifications but suffer from lower throughput and longer production cycles.
Cost Analysis and Resource Requirements
Cell-free protein synthesis typically incurs higher reagent costs due to the need for purified enzymes and substrates, whereas in vivo synthesis leverages cellular machinery, reducing direct expenses but increasing infrastructure and maintenance costs. Resource requirements for cell-free systems emphasize controlled reaction conditions and short duration, enabling rapid prototyping and scalability, while in vivo synthesis demands complex fermentation setups and longer production times. Cost efficiency depends on production scale and application, with cell-free favored for small-scale, high-value proteins and in vivo preferred for bulk protein manufacturing.
Future Trends in Protein Synthesis Technologies
Cell-free protein synthesis technology continues to advance with enhancements in yield, speed, and system scalability, enabling rapid prototyping and complex protein production that traditional in vivo systems often cannot achieve. Emerging trends include integration with microfluidic platforms and artificial intelligence-driven design, enhancing efficiency and automation in protein engineering. Continued development in synthetic biology promises hybrid approaches that combine cell-free systems' flexibility with in vivo systems' complexity for groundbreaking applications in therapeutics and industrial biotechnology.
Transcription-translation coupling
Cell-free protein synthesis enables direct transcription-translation coupling in a controlled environment, bypassing cellular compartmentalization that limits the simultaneous processes seen in vivo protein synthesis.
Lysate-based systems
Lysate-based cell-free protein synthesis systems enable rapid, controlled protein production by utilizing cellular extracts without living cells, offering advantages in speed and scalability compared to traditional in vivo protein synthesis.
Codon optimization
Codon optimization enhances protein yield by matching codon usage to host tRNA abundance in vivo, while cell-free protein synthesis allows flexible codon reprogramming for rapid, high-yield expression of proteins with noncanonical amino acids.
Chaperone-assisted folding
Cell-free protein synthesis enables precise control over chaperone-assisted folding, enhancing protein yield and functionality compared to the inherently complex chaperone networks in in vivo protein synthesis.
Post-translational modification
Cell-free protein synthesis allows precise control and incorporation of non-natural post-translational modifications, unlike in vivo protein synthesis which relies on cellular machinery and may have limited modification scope.
Energy regeneration system
Cell-free protein synthesis utilizes efficient energy regeneration systems like phosphoenolpyruvate or creatine phosphate to sustain protein production outside living cells, whereas in vivo protein synthesis depends on cellular metabolic pathways such as glycolysis and oxidative phosphorylation for energy supply.
Recoding (genetic code expansion)
Cell-free protein synthesis enables more efficient recoding and genetic code expansion by allowing direct manipulation of translational machinery and incorporation of noncanonical amino acids, surpassing the limitations of in vivo protein synthesis.
Plasmid-based expression
Plasmid-based expression in cell-free protein synthesis enables rapid protein production without cell viability constraints, contrasting with in vivo systems where plasmid maintenance and cellular metabolism affect yield and expression efficiency.
Toxic protein expression
Cell-free protein synthesis enables efficient production of toxic proteins by bypassing cellular toxicity constraints inherent in in vivo protein synthesis systems.
Orthogonal translation system
Cell-free protein synthesis enables precise manipulation of the orthogonal translation system for incorporating noncanonical amino acids, offering greater control compared to the complexity and regulatory constraints of in vivo protein synthesis.
Cell-free protein synthesis vs In vivo protein synthesis Infographic
 njnir.com
njnir.com