Recombinase technology offers precise genome editing by catalyzing site-specific DNA rearrangements, ensuring high specificity and minimal off-target effects. Zinc finger nucleases (ZFNs) employ engineered DNA-binding domains coupled with nucleases to induce targeted double-strand breaks, facilitating gene disruption or correction but with a higher risk of unintended mutations. Comparing both, recombinase technology excels in predictable DNA modifications, whereas ZFNs provide broader targeting capabilities albeit with increased complexity in design and potential genomic instability.
Table of Comparison
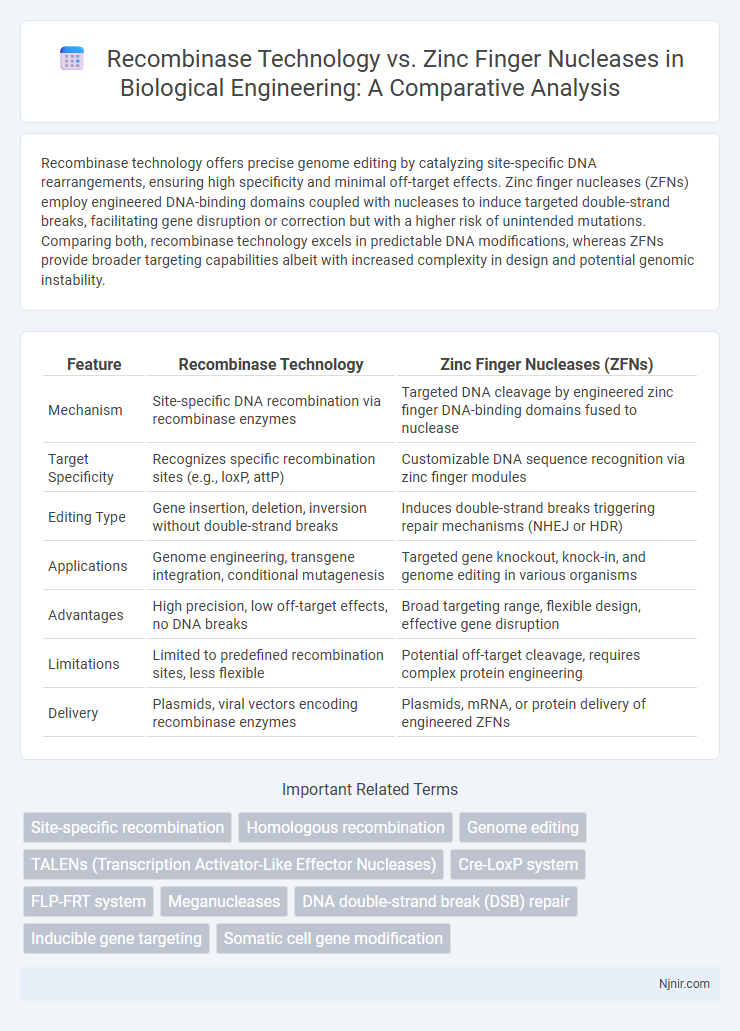
| Feature | Recombinase Technology | Zinc Finger Nucleases (ZFNs) |
|---|---|---|
| Mechanism | Site-specific DNA recombination via recombinase enzymes | Targeted DNA cleavage by engineered zinc finger DNA-binding domains fused to nuclease |
| Target Specificity | Recognizes specific recombination sites (e.g., loxP, attP) | Customizable DNA sequence recognition via zinc finger modules |
| Editing Type | Gene insertion, deletion, inversion without double-strand breaks | Induces double-strand breaks triggering repair mechanisms (NHEJ or HDR) |
| Applications | Genome engineering, transgene integration, conditional mutagenesis | Targeted gene knockout, knock-in, and genome editing in various organisms |
| Advantages | High precision, low off-target effects, no DNA breaks | Broad targeting range, flexible design, effective gene disruption |
| Limitations | Limited to predefined recombination sites, less flexible | Potential off-target cleavage, requires complex protein engineering |
| Delivery | Plasmids, viral vectors encoding recombinase enzymes | Plasmids, mRNA, or protein delivery of engineered ZFNs |
Overview of Recombinase Technology and Zinc Finger Nucleases
Recombinase technology utilizes site-specific recombinases like Cre or FLP enzymes to catalyze precise DNA rearrangements, enabling targeted gene insertion, deletion, or inversion in genomes. Zinc finger nucleases (ZFNs) consist of engineered zinc finger DNA-binding domains fused to the FokI nuclease, facilitating double-strand breaks at defined genomic loci that enable targeted gene editing through cellular repair mechanisms. While recombinases mediate sequence-specific recombination without DNA cleavage, ZFNs introduce targeted double-strand breaks to promote gene modification, highlighting distinct mechanisms in genome engineering applications.
Mechanisms of Genome Modification
Recombinase technology modifies genomes through site-specific recombination by recognizing and cutting DNA at particular sequences, enabling precise insertion or deletion of genetic material. Zinc finger nucleases (ZFNs) utilize engineered DNA-binding domains combined with a nuclease to induce double-strand breaks at targeted genomic loci, triggering cellular repair pathways such as non-homologous end joining or homology-directed repair for targeted gene editing. While recombinases operate via catalyzing recombination at defined sites, ZFNs rely on programmable DNA cleavage and subsequent endogenous repair mechanisms for genome modification.
Specificity and Targeting Efficiency
Recombinase technology offers high specificity by recognizing specific DNA sequences for precise genetic modifications, whereas zinc finger nucleases (ZFNs) combine DNA-binding domains with a nuclease to create targeted double-strand breaks, enhancing targeting efficiency. ZFNs provide greater flexibility in targeting diverse genomic loci due to customizable zinc finger arrays, while recombinase systems are limited to pre-defined target sites. The efficiency of ZFNs in inducing site-specific mutations often surpasses that of recombinases, making ZFNs more suitable for applications requiring precise genome editing with high targeting efficiency.
Applications in Genetic Engineering
Recombinase technology enables precise DNA insertion, deletion, or inversion within genomes, making it highly effective for creating transgenic animals and conditional gene knockouts. Zinc finger nucleases (ZFNs) facilitate targeted genome editing by inducing double-strand breaks at specific DNA sequences, widely used in gene therapy, crop improvement, and functional genomics. Both methods contribute to genetic engineering, with recombinases offering site-specific recombination and ZFNs providing custom gene disruption or correction capabilities.
Advantages of Recombinase Systems
Recombinase technology offers precise site-specific DNA integration without inducing double-strand breaks, significantly reducing off-target effects compared to Zinc Finger Nucleases (ZFNs). Its ability to mediate seamless DNA modifications simplifies gene editing workflows and enhances genomic stability. High target specificity and ease of engineering make recombinase systems advantageous for therapeutic gene insertion and complex genetic rearrangement applications.
Strengths of Zinc Finger Nucleases
Zinc Finger Nucleases (ZFNs) offer precise genome editing capabilities through customizable DNA-binding domains paired with a nuclease, enabling targeted double-strand breaks for efficient gene modification. Their ability to target virtually any genomic sequence with high specificity enhances gene therapy and functional genomics research. ZFNs also exhibit robust performance in complex genomes, providing versatility in therapeutic applications compared to Recombinase technology.
Limitations and Challenges
Recombinase technology faces limitations such as restricted target site specificity and lower efficiency in complex genomes, which hampers its broader application in gene editing. Zinc finger nucleases (ZFNs) present challenges including off-target cleavage, cytotoxicity, and the complexity of designing and engineering specific zinc finger domains for each target sequence. Both technologies require improvements in precision and delivery methods to enhance their therapeutic and research potential.
Comparative Analysis in Therapeutic Uses
Recombinase technology offers site-specific DNA modification with high precision and reduced off-target effects, making it suitable for gene therapy targeting monogenic disorders. Zinc finger nucleases (ZFNs) provide versatile and programmable genome editing capabilities but pose higher risks of off-target cleavage and potential cytotoxicity, which can complicate therapeutic applications. Comparative analyses indicate that recombinase systems excel in safety profiles, while ZFNs demonstrate broader adaptability in targeting diverse genomic loci for therapeutic interventions.
Innovations and Hybrid Approaches
Recombinase technology offers precise site-specific DNA modifications by leveraging engineered enzymes, while Zinc finger nucleases (ZFNs) enable targeted genome editing through customizable DNA-binding domains combined with nuclease activity. Innovations have led to hybrid approaches that integrate recombinase specificity with ZFN-induced double-strand breaks, enhancing editing efficiency and reducing off-target effects. This convergence accelerates the development of versatile gene therapies and functional genomics tools with improved precision and safety profiles.
Future Prospects in Biological Engineering
Recombinase technology offers precise DNA rearrangement capabilities with broad applications in genome editing and synthetic biology, enhancing cell lineage tracing and gene therapy. Zinc finger nucleases (ZFNs) provide targeted gene disruption through programmable DNA cleavage, advancing personalized medicine and functional genomics. Future prospects emphasize combining recombinase specificity with ZFN efficiency to improve gene-editing accuracy, reduce off-target effects, and enable complex genomic modifications in biological engineering.
Site-specific recombination
Recombinase technology enables precise site-specific recombination by recognizing specific DNA sequences, whereas Zinc Finger Nucleases induce targeted double-strand breaks to facilitate genome editing through error-prone repair mechanisms.
Homologous recombination
Recombinase technology enables precise homologous recombination by catalyzing site-specific DNA rearrangements, while zinc finger nucleases induce targeted double-strand breaks to facilitate homologous recombination and gene editing with high specificity.
Genome editing
Recombinase technology enables precise site-specific genome editing by catalyzing DNA rearrangements, while Zinc Finger Nucleases (ZFNs) facilitate targeted genome modifications through engineered DNA cleavage and subsequent repair mechanisms.
TALENs (Transcription Activator-Like Effector Nucleases)
TALENs (Transcription Activator-Like Effector Nucleases) offer higher specificity and efficiency in genome editing compared to recombinase technology and Zinc Finger Nucleases (ZFNs) by utilizing customizable DNA-binding domains for targeted double-strand breaks.
Cre-LoxP system
The Cre-LoxP system, a precise recombinase technology, enables targeted DNA recombination with higher specificity and fewer off-target effects compared to Zinc Finger Nucleases, making it ideal for conditional gene editing in complex genomes.
FLP-FRT system
The FLP-FRT recombinase technology enables precise site-specific DNA recombination with higher accuracy and lower off-target effects compared to Zinc Finger Nucleases, making it ideal for genetic engineering applications requiring targeted genomic modifications.
Meganucleases
Meganucleases offer higher specificity and lower off-target effects compared to Zinc Finger Nucleases and are a key alternative in recombinase technology for precise genome editing.
DNA double-strand break (DSB) repair
Recombinase technology facilitates site-specific DNA double-strand break (DSB) repair through precise strand exchange without causing DSBs, whereas zinc finger nucleases induce targeted DSBs that stimulate cellular repair pathways like non-homologous end joining (NHEJ) or homology-directed repair (HDR) for genome editing.
Inducible gene targeting
Inducible gene targeting using recombinase technology offers precise, reversible control of gene expression compared to the permanent, sequence-specific DNA cleavage by zinc finger nucleases.
Somatic cell gene modification
Recombinase technology enables precise site-specific somatic cell gene modification with lower off-target effects compared to Zinc Finger Nucleases, which offer versatile but potentially higher off-target cleavage activity in genome editing applications.
Recombinase technology vs Zinc finger nucleases Infographic
 njnir.com
njnir.com