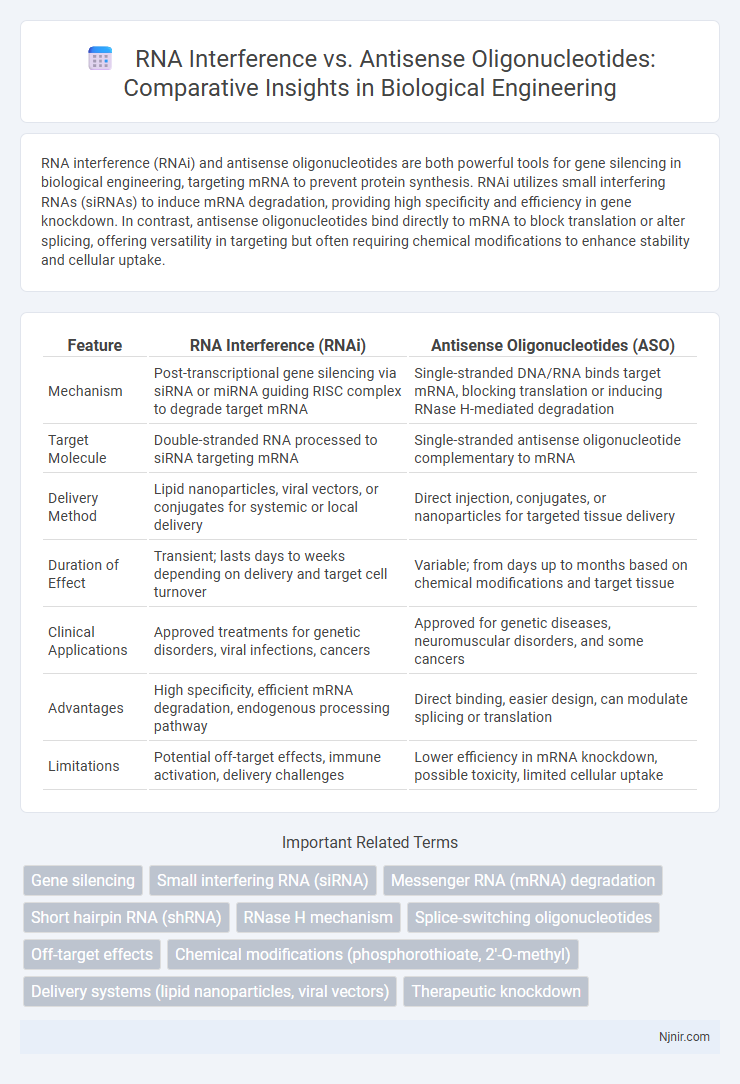
RNA interference (RNAi) and antisense oligonucleotides are both powerful tools for gene silencing in biological engineering, targeting mRNA to prevent protein synthesis. RNAi utilizes small interfering RNAs (siRNAs) to induce mRNA degradation, providing high specificity and efficiency in gene knockdown. In contrast, antisense oligonucleotides bind directly to mRNA to block translation or alter splicing, offering versatility in targeting but often requiring chemical modifications to enhance stability and cellular uptake.
Table of Comparison
| Feature | RNA Interference (RNAi) | Antisense Oligonucleotides (ASO) |
|---|---|---|
| Mechanism | Post-transcriptional gene silencing via siRNA or miRNA guiding RISC complex to degrade target mRNA | Single-stranded DNA/RNA binds target mRNA, blocking translation or inducing RNase H-mediated degradation |
| Target Molecule | Double-stranded RNA processed to siRNA targeting mRNA | Single-stranded antisense oligonucleotide complementary to mRNA |
| Delivery Method | Lipid nanoparticles, viral vectors, or conjugates for systemic or local delivery | Direct injection, conjugates, or nanoparticles for targeted tissue delivery |
| Duration of Effect | Transient; lasts days to weeks depending on delivery and target cell turnover | Variable; from days up to months based on chemical modifications and target tissue |
| Clinical Applications | Approved treatments for genetic disorders, viral infections, cancers | Approved for genetic diseases, neuromuscular disorders, and some cancers |
| Advantages | High specificity, efficient mRNA degradation, endogenous processing pathway | Direct binding, easier design, can modulate splicing or translation |
| Limitations | Potential off-target effects, immune activation, delivery challenges | Lower efficiency in mRNA knockdown, possible toxicity, limited cellular uptake |
Introduction to Gene Silencing Technologies
RNA interference (RNAi) and antisense oligonucleotides (ASOs) are two prominent gene silencing technologies that regulate gene expression at the RNA level. RNAi utilizes small interfering RNA (siRNA) molecules to target and degrade specific mRNA sequences, leading to post-transcriptional gene silencing, whereas ASOs are short, single-stranded DNA or RNA molecules that bind complementary mRNA to block translation or induce RNase H-mediated degradation. Both technologies offer precise and sequence-specific modulation of gene activity, making them powerful tools for therapeutic development and functional genomics research.
Mechanistic Overview: RNA Interference vs Antisense Oligonucleotides
RNA interference (RNAi) functions by utilizing small interfering RNAs (siRNAs) that guide the RNA-induced silencing complex (RISC) to degrade complementary mRNA, thereby preventing translation and reducing gene expression. Antisense oligonucleotides (ASOs) operate by hybridizing directly to target mRNA sequences, modulating splicing, promoting RNase H-mediated degradation, or blocking translation without involving RISC. While RNAi relies on endogenous cellular pathways with siRNA incorporation, ASOs provide versatile mechanisms for gene regulation through direct base pairing and enzymatic recruitment.
Molecular Components and Design Strategies
RNA interference (RNAi) relies on small interfering RNAs (siRNAs) or microRNAs (miRNAs) that guide the RNA-induced silencing complex (RISC) to complementary mRNA for cleavage, whereas antisense oligonucleotides (ASOs) are single-stranded DNA or RNA molecules that bind directly to target mRNA to modulate splicing or induce RNase H-mediated degradation. RNAi design strategies emphasize selecting siRNA sequences with optimal thermodynamic stability and minimal off-target effects, including chemical modifications to enhance stability and reduce immunogenicity. ASO design focuses on backbone chemistry modifications such as phosphorothioate linkages and 2'-O-methyl or locked nucleic acid (LNA) bases to improve binding affinity, nuclease resistance, and target specificity.
Target Recognition and Binding Specificity
RNA interference (RNAi) utilizes small interfering RNA (siRNA) molecules that recognize and bind complementary mRNA sequences with high specificity through base pairing, inducing mRNA cleavage and degradation. In contrast, antisense oligonucleotides (ASOs) typically bind target RNA via Watson-Crick base pairing but exhibit variable specificity influenced by chemical modifications and local RNA structure, often leading to inhibition of translation or alteration of splicing. RNAi generally achieves more robust, sequence-specific gene silencing due to incorporation into the RNA-induced silencing complex (RISC), while ASOs offer versatile target engagement mechanisms depending on design and chemistry.
Delivery Systems in Biological Engineering
RNA interference (RNAi) and antisense oligonucleotides (ASOs) rely heavily on advanced delivery systems such as lipid nanoparticles, viral vectors, and polymer-based carriers to traverse cellular membranes and achieve targeted gene silencing in biological engineering applications. The efficiency of RNAi delivery is often enhanced by small interfering RNA (siRNA) encapsulation in lipid-based nanoparticles, promoting endosomal escape and cytoplasmic release, whereas ASOs benefit from chemical modifications improving stability and cellular uptake through conjugation with cell-penetrating peptides or receptor-targeting ligands. Optimization of these delivery systems addresses challenges like nuclease degradation, off-target effects, and immune activation, crucial for therapeutic efficacy in gene regulation and disease treatment.
Efficacy and Durability of Gene Silencing
RNA interference (RNAi) typically achieves higher efficacy in gene silencing through sequence-specific degradation of mRNA, leading to robust and rapid knockdown of target gene expression. Antisense oligonucleotides (ASOs) often provide more durable gene silencing by promoting RNase H-mediated cleavage or modulating splicing, thereby sustaining target inhibition over extended periods. The choice between RNAi and ASOs depends on the desired balance between immediate potency and long-term gene silencing stability in therapeutic applications.
Off-Target Effects and Specificity Challenges
RNA interference (RNAi) and antisense oligonucleotides (ASOs) both face off-target effects due to partial sequence complementarity, leading to unintended gene silencing. RNAi utilizes the RNA-induced silencing complex (RISC) and can inadvertently target multiple mRNAs with similar sequences, complicating specificity. ASOs rely on direct hybridization to target mRNA but may bind non-specifically to related transcripts or secondary structures, posing challenges in achieving precise gene modulation.
Therapeutic Applications: Comparative Advances
RNA interference (RNAi) and antisense oligonucleotides (ASOs) both offer targeted gene silencing but differ in mechanism and therapeutic scope. RNAi utilizes small interfering RNAs (siRNAs) to induce mRNA degradation, demonstrating efficacy in diseases like hereditary transthyretin amyloidosis and acute hepatic porphyria. ASOs act by binding complementary mRNA sequences to modulate splicing or block translation, showing significant advances in treating spinal muscular atrophy and Duchenne muscular dystrophy with FDA-approved drugs such as nusinersen and eteplirsen.
Current Limitations and Innovation Pathways
RNA interference (RNAi) faces challenges including off-target effects, immune activation, and delivery barriers to specific tissues, while antisense oligonucleotides (ASOs) are limited by stability, cellular uptake, and potential toxicity. Innovations in lipid nanoparticle carriers, chemical modifications like 2'-O-methyl and locked nucleic acids (LNAs), and targeted conjugates aim to enhance stability, specificity, and reduce immunogenicity in both RNAi and ASO therapeutics. Emerging strategies focus on improving endosomal escape mechanisms and leveraging artificial intelligence for optimized sequence design to overcome current delivery and efficacy constraints.
Future Directions in RNA-Based Modulation
RNA interference (RNAi) and antisense oligonucleotides (ASOs) both serve as powerful tools for gene silencing, with RNAi utilizing small interfering RNAs (siRNAs) and ASOs employing single-stranded DNA or RNA to modulate mRNA function. Future directions in RNA-based modulation emphasize enhancing delivery mechanisms, increasing target specificity, and minimizing off-target effects to treat genetic disorders, cancers, and viral infections more effectively. Emerging technologies such as CRISPR-RNAi hybrids and chemically modified oligonucleotides are expanding the therapeutic potential and precision of these modalities in personalized medicine.
Gene silencing
RNA interference (RNAi) employs siRNA or miRNA molecules to induce sequence-specific degradation of target mRNA, enabling potent and reversible gene silencing, while antisense oligonucleotides (ASOs) bind complementary RNA to modulate splicing or block translation, offering versatile but often less efficient gene silencing options.
Small interfering RNA (siRNA)
Small interfering RNA (siRNA) in RNA interference (RNAi) efficiently silences gene expression by targeting mRNA for degradation, offering higher specificity and potency compared to antisense oligonucleotides.
Messenger RNA (mRNA) degradation
RNA interference (RNAi) efficiently induces messenger RNA (mRNA) degradation through the RNA-induced silencing complex (RISC), whereas antisense oligonucleotides (ASOs) promote mRNA cleavage primarily via RNase H activation.
Short hairpin RNA (shRNA)
Short hairpin RNA (shRNA) used in RNA interference (RNAi) efficiently silences target gene expression by inducing sequence-specific mRNA degradation, offering longer-lasting effects compared to antisense oligonucleotides that bind mRNA but typically exhibit transient and less stable gene knockdown.
RNase H mechanism
RNA interference (RNAi) primarily uses the RNA-induced silencing complex (RISC) to degrade target mRNA, while antisense oligonucleotides (ASOs) recruit RNase H to cleave RNA-DNA hybrids, directly exploiting the RNase H mechanism for gene silencing.
Splice-switching oligonucleotides
Splice-switching oligonucleotides (SSOs) are a specialized class of antisense oligonucleotides designed to modulate pre-mRNA splicing by binding to splice sites, providing precise gene expression regulation distinct from RNA interference (RNAi), which primarily mediates mRNA degradation through the RNA-induced silencing complex.
Off-target effects
RNA interference (RNAi) exhibits higher off-target effects due to microRNA-like seed region binding, whereas antisense oligonucleotides (ASOs) tend to have more sequence-specific off-target interactions, resulting in fewer unintended gene silencing events.
Chemical modifications (phosphorothioate, 2'-O-methyl)
Phosphorothioate and 2'-O-methyl chemical modifications enhance stability and reduce off-target effects in RNA interference (RNAi) and antisense oligonucleotides, respectively, optimizing their therapeutic efficacy.
Delivery systems (lipid nanoparticles, viral vectors)
Lipid nanoparticles enable efficient cytoplasmic delivery of RNA interference (RNAi) molecules, while viral vectors provide robust nuclear delivery of antisense oligonucleotides, optimizing therapeutic gene silencing.
Therapeutic knockdown
RNA interference (RNAi) achieves therapeutic knockdown by utilizing small interfering RNAs (siRNAs) to degrade target mRNA with high specificity, whereas antisense oligonucleotides (ASOs) bind complementary mRNA sequences to modulate splicing or induce RNase H-mediated cleavage, each offering distinct mechanisms and delivery challenges for gene silencing.
RNA interference (RNAi) vs Antisense oligonucleotides Infographic
 njnir.com
njnir.com