Digital PCR offers absolute quantification of nucleic acids by partitioning the sample into thousands of individual reactions, enhancing sensitivity and precision compared to qPCR. Unlike qPCR, which relies on amplification curves and standard curves for relative quantification, digital PCR allows direct counting of target DNA molecules, reducing amplification bias. This makes digital PCR particularly valuable for detecting low-abundance targets, rare mutations, and copy number variations in biomedical research and clinical diagnostics.
Table of Comparison
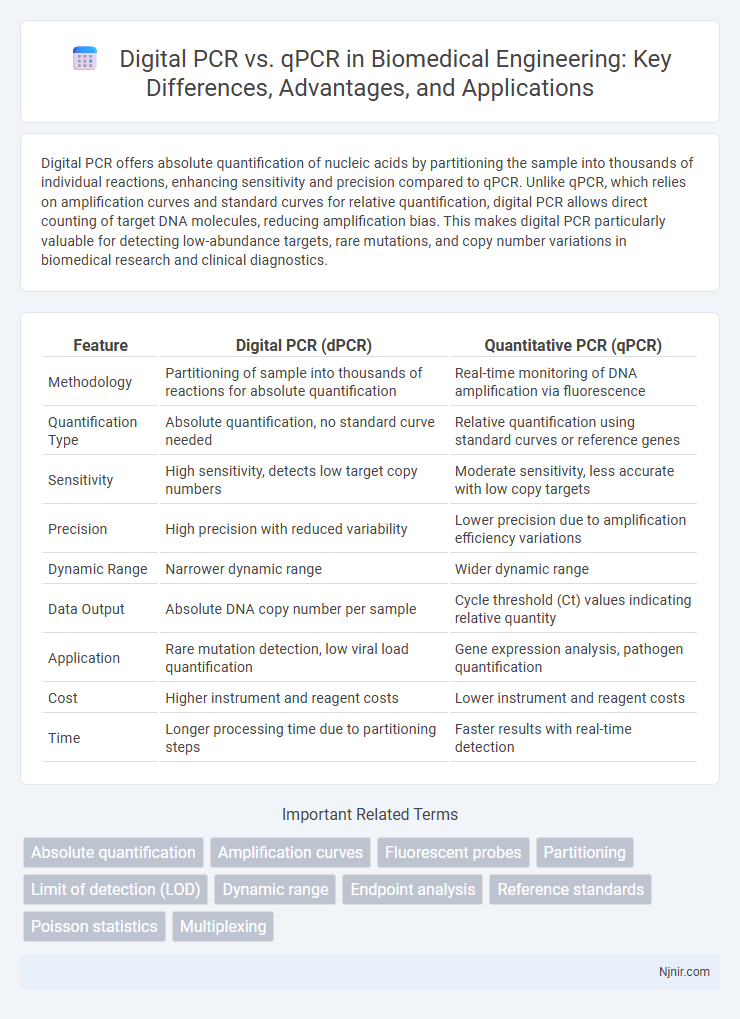
| Feature | Digital PCR (dPCR) | Quantitative PCR (qPCR) |
|---|---|---|
| Methodology | Partitioning of sample into thousands of reactions for absolute quantification | Real-time monitoring of DNA amplification via fluorescence |
| Quantification Type | Absolute quantification, no standard curve needed | Relative quantification using standard curves or reference genes |
| Sensitivity | High sensitivity, detects low target copy numbers | Moderate sensitivity, less accurate with low copy targets |
| Precision | High precision with reduced variability | Lower precision due to amplification efficiency variations |
| Dynamic Range | Narrower dynamic range | Wider dynamic range |
| Data Output | Absolute DNA copy number per sample | Cycle threshold (Ct) values indicating relative quantity |
| Application | Rare mutation detection, low viral load quantification | Gene expression analysis, pathogen quantification |
| Cost | Higher instrument and reagent costs | Lower instrument and reagent costs |
| Time | Longer processing time due to partitioning steps | Faster results with real-time detection |
Introduction to Digital PCR and qPCR Technologies
Digital PCR (dPCR) partitions a sample into thousands of individual reactions to provide absolute quantification of nucleic acids without the need for standard curves, enhancing sensitivity and precision. qPCR (quantitative PCR) measures DNA amplification in real-time by monitoring fluorescence intensity, allowing relative quantification through cycle threshold (Ct) values. Both technologies rely on PCR amplification but differ in data output, with dPCR offering higher accuracy for low target concentrations and qPCR enabling rapid and high-throughput analysis.
Principles and Workflow of qPCR
qPCR, or quantitative polymerase chain reaction, relies on the exponential amplification of target DNA sequences combined with fluorescent probes or dyes to quantify nucleic acids in real-time during each amplification cycle. The workflow includes sample preparation, reverse transcription if starting from RNA, amplification using sequence-specific primers, and fluorescent signal detection by a thermocycler equipped with an optical system. This method allows precise quantification of gene expression based on threshold cycle (Ct) values, reflecting the initial amount of target nucleic acid in the sample.
Digital PCR: Fundamentals and Methodology
Digital PCR (dPCR) offers precise quantification of nucleic acids by partitioning a sample into thousands of individual reactions, enabling absolute measurement without the need for standard curves. This methodology amplifies DNA in discrete partitions, allowing detection of rare variants and low-abundance targets with superior sensitivity and accuracy compared to quantitative PCR (qPCR). dPCR's fundamental principle relies on Poisson statistics to determine the exact number of target molecules, providing robust results in diagnostic and research applications.
Sensitivity and Specificity Comparison
Digital PCR (dPCR) offers superior sensitivity compared to qPCR by enabling absolute quantification of low-abundance targets without reliance on standard curves, which reduces quantification bias. The partitioning of samples in dPCR enhances specificity by minimizing the impact of inhibitors and background noise, resulting in more accurate detection of rare mutations or low-level nucleic acid variants. In contrast, qPCR's sensitivity and specificity can be limited by amplification efficiency and primer-dimer artifacts, affecting precise quantification in samples with complex backgrounds.
Quantification Accuracy: Absolute vs Relative Methods
Digital PCR (dPCR) offers absolute quantification by partitioning samples into thousands of individual reactions, allowing direct counting of DNA molecules without reliance on standard curves. In contrast, quantitative PCR (qPCR) provides relative quantification based on fluorescent signal intensity compared to reference genes or standards, which can introduce variability and limit precision. The absolute quantification capability of dPCR enhances sensitivity and reproducibility, making it the preferred method for applications requiring precise copy number determination.
Throughput and Sample Requirements
Digital PCR offers higher throughput by enabling simultaneous partitioning of thousands of individual reactions, which allows precise quantification of low-abundance targets in complex samples. In contrast, qPCR typically processes fewer samples per run and requires larger sample volumes to achieve reliable quantification, limiting its efficiency for high-throughput applications. Sample requirements for digital PCR are minimal, often in the range of picograms to nanograms of DNA, while qPCR usually demands higher input quantities to maintain sensitivity and accuracy.
Detection of Rare Mutations and Low-Abundance Targets
Digital PCR (dPCR) offers superior sensitivity for detecting rare mutations and low-abundance targets compared to quantitative PCR (qPCR) by partitioning the sample into thousands of individual reactions, enabling precise quantification of minute nucleic acid amounts. Unlike qPCR, which relies on amplification curves and relative quantification, dPCR provides absolute quantification without standards, enhancing detection accuracy in challenging samples such as liquid biopsies and heterogeneous tumor tissues. This enhanced sensitivity and precision make dPCR the preferred method for applications requiring rare mutation detection, minimal residual disease monitoring, and viral load quantification.
Applications in Biomedical Research and Diagnostics
Digital PCR (dPCR) offers absolute quantification of nucleic acids, enhancing sensitivity and precision in detecting low-abundance targets, crucial for rare mutation analysis and viral load monitoring in biomedical research. Quantitative PCR (qPCR) remains widely used for gene expression profiling, pathogen detection, and genotyping due to its high throughput and rapid results. In diagnostics, dPCR improves accuracy in minimal residual disease detection and liquid biopsy applications, while qPCR provides robust screening tools for infectious diseases and genetic disorders.
Cost, Equipment, and Practical Considerations
Digital PCR (dPCR) typically incurs higher costs due to specialized equipment such as microfluidic partitioning systems and fluorescence detectors, while qPCR machines are more affordable and widely available in most laboratories. The complexity of dPCR equipment requires skilled operators and longer setup times, whereas qPCR offers faster turnaround with simpler workflows and well-established protocols. Practical considerations favor qPCR for routine, high-throughput applications, while dPCR excels in absolute quantification and detecting low-abundance targets despite increased operational expenses.
Future Trends in PCR-Based Molecular Analysis
Digital PCR (dPCR) offers enhanced precision in absolute quantification and rare mutation detection, driving its adoption in clinical diagnostics and liquid biopsy applications. qPCR remains widely used due to its high throughput and ease of integration but is evolving with multiplexing and real-time data analytics advancements. Future trends emphasize integrating dPCR with microfluidics and artificial intelligence to improve sensitivity, reduce assay time, and enable personalized medicine through highly accurate molecular analysis.
Absolute quantification
Digital PCR provides precise absolute quantification by partitioning samples into thousands of micro-reactions, unlike qPCR which relies on standard curves for relative quantification.
Amplification curves
Digital PCR produces discrete amplification signals for absolute quantification, while qPCR generates continuous amplification curves reflecting relative DNA concentration through cycle threshold values.
Fluorescent probes
Digital PCR employs partitioned reactions with fluorescent probes for absolute quantification, whereas qPCR uses fluorescent probes for relative quantification based on amplification cycles.
Partitioning
Digital PCR achieves precise quantification by partitioning samples into thousands of nanoliter-sized reactions, whereas qPCR amplifies DNA in bulk without sample partitioning.
Limit of detection (LOD)
Digital PCR offers a lower limit of detection (LOD) than qPCR by enabling absolute quantification of low-abundance DNA targets with higher sensitivity and precision.
Dynamic range
Digital PCR offers a broader dynamic range with higher sensitivity and absolute quantification compared to qPCR, which is limited by its reliance on standard curves and amplification efficiency.
Endpoint analysis
Digital PCR provides absolute quantification through partitioned endpoint analysis, offering higher precision and sensitivity compared to qPCR's relative quantification based on real-time amplification curves.
Reference standards
Digital PCR provides absolute quantification without the need for reference standards, whereas qPCR relies on reference standards for relative quantification accuracy.
Poisson statistics
Digital PCR utilizes Poisson statistics to precisely quantify nucleic acid copies by partitioning samples into thousands of individual reactions, unlike qPCR which measures amplification in bulk and relies on standard curves.
Multiplexing
Digital PCR enables higher multiplexing capabilities than qPCR by partitioning samples into thousands of reactions, allowing simultaneous quantification of multiple targets with greater precision and sensitivity.
Digital PCR vs qPCR Infographic
 njnir.com
njnir.com