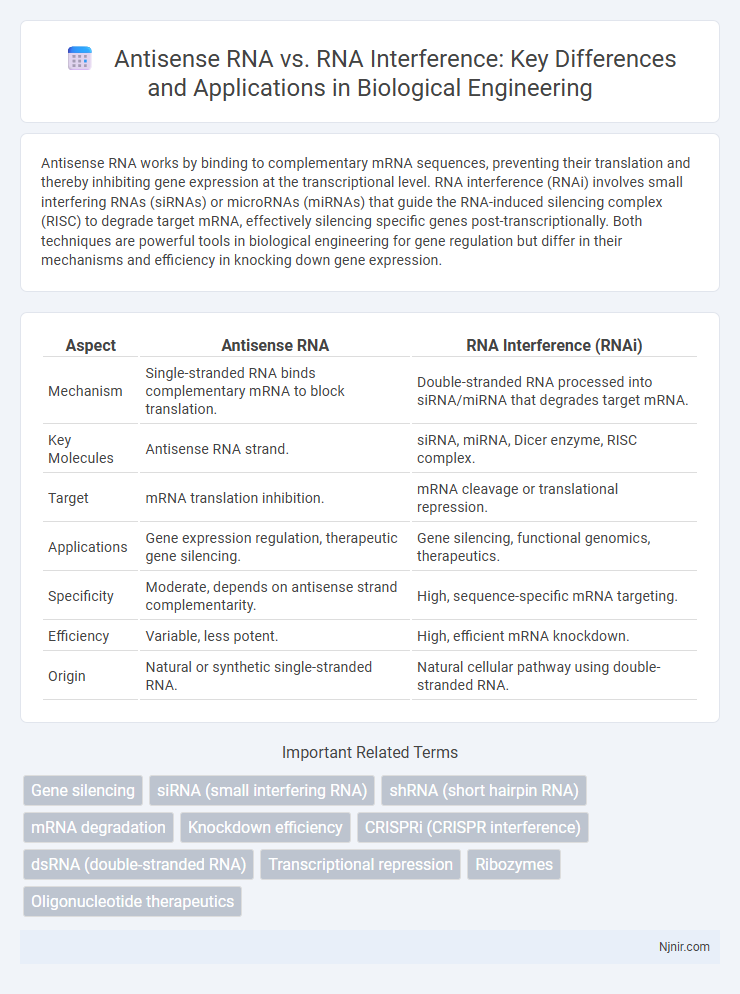
Antisense RNA works by binding to complementary mRNA sequences, preventing their translation and thereby inhibiting gene expression at the transcriptional level. RNA interference (RNAi) involves small interfering RNAs (siRNAs) or microRNAs (miRNAs) that guide the RNA-induced silencing complex (RISC) to degrade target mRNA, effectively silencing specific genes post-transcriptionally. Both techniques are powerful tools in biological engineering for gene regulation but differ in their mechanisms and efficiency in knocking down gene expression.
Table of Comparison
| Aspect | Antisense RNA | RNA Interference (RNAi) |
|---|---|---|
| Mechanism | Single-stranded RNA binds complementary mRNA to block translation. | Double-stranded RNA processed into siRNA/miRNA that degrades target mRNA. |
| Key Molecules | Antisense RNA strand. | siRNA, miRNA, Dicer enzyme, RISC complex. |
| Target | mRNA translation inhibition. | mRNA cleavage or translational repression. |
| Applications | Gene expression regulation, therapeutic gene silencing. | Gene silencing, functional genomics, therapeutics. |
| Specificity | Moderate, depends on antisense strand complementarity. | High, sequence-specific mRNA targeting. |
| Efficiency | Variable, less potent. | High, efficient mRNA knockdown. |
| Origin | Natural or synthetic single-stranded RNA. | Natural cellular pathway using double-stranded RNA. |
Introduction to Gene Silencing Mechanisms
Gene silencing mechanisms involve the regulation of gene expression by preventing the translation of specific mRNA molecules. Antisense RNA works by binding complementary mRNA sequences to block translation directly, while RNA interference (RNAi) employs small interfering RNAs (siRNAs) or microRNAs (miRNAs) to induce mRNA degradation or translational repression through the RNA-induced silencing complex (RISC). Both methods are crucial for studying gene function and developing therapeutic strategies for genetic disorders and viral infections.
Overview of Antisense RNA Technology
Antisense RNA technology utilizes short, single-stranded RNA molecules designed to bind specifically to target mRNA, blocking translation and gene expression. This approach enables precise downregulation of disease-related genes by preventing protein synthesis at the post-transcriptional level. Unlike RNA interference, which relies on the cellular RNA-induced silencing complex (RISC), antisense RNA operates through direct hybridization with complementary mRNA sequences.
Fundamentals of RNA Interference (RNAi)
RNA interference (RNAi) is a biological process in which small RNA molecules, such as small interfering RNA (siRNA) or microRNA (miRNA), mediate the sequence-specific degradation or translational repression of target mRNA molecules, effectively silencing gene expression post-transcriptionally. This mechanism involves the incorporation of double-stranded RNA (dsRNA) into the RNA-induced silencing complex (RISC), where the guide strand directs the complex to complementary mRNA for cleavage or inhibition. In contrast, antisense RNA is a single-stranded RNA molecule complementary to a target mRNA that binds and blocks translation without the involvement of the RISC complex or mRNA cleavage, providing a more direct and less enzymatically driven method of gene silencing.
Molecular Mechanisms: Antisense RNA vs RNAi
Antisense RNA exerts its effect by binding complementary mRNA sequences, blocking translation or promoting mRNA degradation through RNase H activity. RNA interference (RNAi) involves small interfering RNAs (siRNAs) or microRNAs (miRNAs) that guide the RNA-induced silencing complex (RISC) to target mRNAs, leading to cleavage or translational repression. While antisense RNA acts primarily through direct base pairing and RNase H recruitment, RNAi depends on specialized protein complexes that mediate gene silencing with higher specificity and efficiency.
Efficiency and Specificity in Gene Regulation
Antisense RNA modulates gene expression by binding complementary mRNA sequences, but its efficiency is often limited by incomplete hybridization and susceptibility to degradation. RNA interference (RNAi) employs small interfering RNAs (siRNAs) or microRNAs (miRNAs) that guide the RNA-induced silencing complex (RISC) to target mRNAs, achieving higher specificity and robust knockdown of gene expression. RNAi's mechanism allows for more precise gene silencing with reduced off-target effects compared to antisense RNA approaches.
Applications in Functional Genomics
Antisense RNA and RNA interference (RNAi) are pivotal in functional genomics for gene expression modulation, where antisense RNA binds complementary mRNA to block translation, while RNAi uses small interfering RNAs (siRNAs) or microRNAs (miRNAs) to trigger mRNA degradation. Antisense RNA finds applications in validating gene function by specific inhibition, often used in prokaryotic and eukaryotic systems for targeted gene knockdown. RNAi offers a more efficient and widespread tool for genome-wide screening, facilitating gene silencing in high-throughput studies to elucidate gene roles in development, disease mechanisms, and drug target discovery.
Therapeutic Potential: Antisense RNA and RNAi
Antisense RNA and RNA interference (RNAi) both offer promising therapeutic potential by specifically targeting mRNA to modulate gene expression and treat genetic disorders. Antisense RNA binds directly to complementary mRNA sequences to block translation and promote degradation, whereas RNAi utilizes small interfering RNAs (siRNAs) to recruit the RNA-induced silencing complex (RISC) for targeted mRNA cleavage. Clinical applications demonstrate RNAi's efficiency in silencing genes in diseases like hereditary transthyretin amyloidosis, while antisense oligonucleotides show benefits in treating spinal muscular atrophy, highlighting their complementary roles in gene therapy.
Delivery Systems and Experimental Challenges
Antisense RNA delivery systems often utilize lipid nanoparticles or viral vectors to enhance cellular uptake and stability, whereas RNA interference (RNAi) typically employs small interfering RNA (siRNA) encapsulated in lipid-based carriers or polymeric nanoparticles for targeted gene silencing. Experimental challenges for antisense RNA include off-target effects and intracellular degradation, while RNAi faces issues such as immune stimulation, delivery efficiency to specific tissues, and endosomal escape. Both approaches require optimization of delivery vehicles to improve bioavailability and minimize cytotoxicity in therapeutic applications.
Comparative Advantages and Limitations
Antisense RNA provides a targeted approach by binding directly to complementary mRNA sequences, effectively blocking translation with high specificity and minimal off-target effects, but its delivery and stability pose significant challenges in vivo. RNA interference (RNAi) employs small interfering RNAs (siRNAs) or microRNAs (miRNAs) to induce mRNA degradation through the RNA-induced silencing complex (RISC), offering potent gene silencing and the ability to target multiple genes simultaneously, yet it risks off-target gene suppression and immune activation. Both techniques require careful design to maximize efficacy and minimize unintended effects, with RNAi generally favored for robust knockdown and antisense RNA preferred when precise, single-gene modulation is critical.
Future Perspectives in Biological Engineering
Future perspectives in biological engineering highlight the potential of antisense RNA and RNA interference (RNAi) for precise gene regulation and therapeutic applications. Advances in delivery systems and molecular targeting enhance the specificity and efficiency of antisense oligonucleotides and small interfering RNAs (siRNAs), enabling treatment of genetic disorders, cancers, and viral infections. Integration with CRISPR technology and synthetic biology may further optimize RNA-based gene silencing strategies, driving innovation in personalized medicine and functional genomics.
Gene silencing
Antisense RNA silences genes by binding complementary mRNA sequences to block translation, whereas RNA interference employs small interfering RNAs (siRNAs) or microRNAs (miRNAs) to induce mRNA degradation or translational repression for gene silencing.
siRNA (small interfering RNA)
siRNA (small interfering RNA) mediates RNA interference by guiding the RNA-induced silencing complex (RISC) to degrade complementary mRNA, whereas antisense RNA functions by directly binding to target mRNA to block translation without promoting degradation.
shRNA (short hairpin RNA)
shRNA (short hairpin RNA) mediates RNA interference by forming a hairpin structure processed into siRNA, enabling efficient and stable gene silencing compared to antisense RNA mechanisms that rely on direct sequence hybridization to block mRNA translation.
mRNA degradation
Antisense RNA induces mRNA degradation by forming complementary duplexes that recruit RNase H, while RNA interference triggers mRNA cleavage through the RISC complex guided by siRNA or miRNA.
Knockdown efficiency
RNA interference typically achieves higher knockdown efficiency than antisense RNA due to its ability to utilize cellular machinery for targeted mRNA degradation.
CRISPRi (CRISPR interference)
CRISPR interference (CRISPRi) utilizes a catalytically dead Cas9 protein guided by sgRNA to specifically block transcription, offering a more precise and efficient gene repression method compared to traditional antisense RNA and RNA interference mechanisms.
dsRNA (double-stranded RNA)
Double-stranded RNA (dsRNA) serves as the key trigger in RNA interference (RNAi) by initiating the degradation of target mRNA, whereas antisense RNA typically involves single-stranded RNA binding to complementary mRNA to block translation.
Transcriptional repression
Antisense RNA mediates transcriptional repression by directly binding to complementary mRNA sequences to block transcription, whereas RNA interference silences gene expression post-transcriptionally through small interfering RNAs that target mRNA degradation.
Ribozymes
Ribozymes are catalytic RNA molecules that cleave target RNA sequences with high specificity, distinguishing them from antisense RNA which blocks translation by base pairing and RNA interference which employs protein complexes like RISC for targeted mRNA degradation.
Oligonucleotide therapeutics
Oligonucleotide therapeutics utilize antisense RNA to bind specific mRNA sequences and inhibit gene expression directly, while RNA interference employs small interfering RNAs (siRNAs) to trigger mRNA degradation through the RNA-induced silencing complex (RISC), offering distinct mechanisms for targeted gene silencing in disease treatment.
Antisense RNA vs RNA interference Infographic
 njnir.com
njnir.com