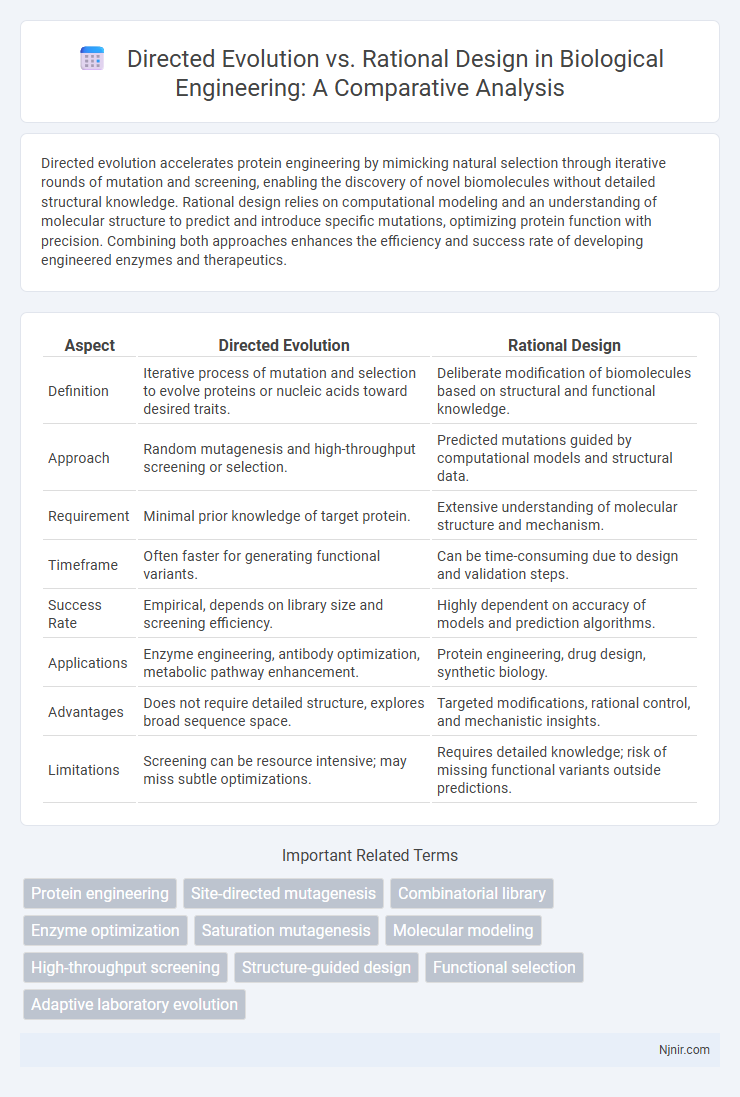
Directed evolution accelerates protein engineering by mimicking natural selection through iterative rounds of mutation and screening, enabling the discovery of novel biomolecules without detailed structural knowledge. Rational design relies on computational modeling and an understanding of molecular structure to predict and introduce specific mutations, optimizing protein function with precision. Combining both approaches enhances the efficiency and success rate of developing engineered enzymes and therapeutics.
Table of Comparison
| Aspect | Directed Evolution | Rational Design |
|---|---|---|
| Definition | Iterative process of mutation and selection to evolve proteins or nucleic acids toward desired traits. | Deliberate modification of biomolecules based on structural and functional knowledge. |
| Approach | Random mutagenesis and high-throughput screening or selection. | Predicted mutations guided by computational models and structural data. |
| Requirement | Minimal prior knowledge of target protein. | Extensive understanding of molecular structure and mechanism. |
| Timeframe | Often faster for generating functional variants. | Can be time-consuming due to design and validation steps. |
| Success Rate | Empirical, depends on library size and screening efficiency. | Highly dependent on accuracy of models and prediction algorithms. |
| Applications | Enzyme engineering, antibody optimization, metabolic pathway enhancement. | Protein engineering, drug design, synthetic biology. |
| Advantages | Does not require detailed structure, explores broad sequence space. | Targeted modifications, rational control, and mechanistic insights. |
| Limitations | Screening can be resource intensive; may miss subtle optimizations. | Requires detailed knowledge; risk of missing functional variants outside predictions. |
Introduction to Protein Engineering Approaches
Directed evolution mimics natural selection by generating random protein variants and selecting those with enhanced functions, enabling rapid optimization without prior structural knowledge. Rational design relies on detailed understanding of protein structure and function, using computational tools to introduce specific mutations for desired traits. Combining these protein engineering approaches accelerates the development of enzymes with improved stability, activity, and specificity for industrial and therapeutic applications.
Overview of Directed Evolution
Directed evolution mimics natural selection to engineer proteins by iterative rounds of mutagenesis and screening, enabling the discovery of biomolecules with enhanced or novel functions. This technique bypasses the need for detailed structural information, making it highly effective for evolving enzymes, antibodies, and other proteins with desired traits. Directed evolution has revolutionized protein engineering by providing a practical approach to optimize complex biological functions through high-throughput experimentation.
Fundamentals of Rational Design
Rational design in protein engineering relies on detailed knowledge of protein structure, function, and mechanism to introduce specific mutations that enhance desired traits. This approach uses computational models, structural biology data, and active site analysis to predict amino acid changes improving enzyme activity, stability, or specificity. Unlike directed evolution, which screens random mutations, rational design offers precise control, reducing experimental iterations by targeting key residues informed by molecular insights.
Comparative Mechanisms: Directed Evolution vs Rational Design
Directed evolution mimics natural selection by generating genetic diversity through random mutations followed by iterative screening to enhance desired protein traits. Rational design relies on detailed structural and computational knowledge to introduce specific mutations aimed at improving function based on predicted molecular interactions. Directed evolution excels in exploring vast sequence space without prior mechanistic understanding, whereas rational design offers targeted modifications grounded in mechanistic insights but is limited by structural information accuracy.
Applications in Enzyme Engineering
Directed evolution accelerates the natural selection process to create enzymes with enhanced stability, activity, or substrate specificity, making it a preferred method for optimizing biocatalysts in industrial processes like biofuel production and pharmaceutical synthesis. Rational design leverages detailed knowledge of enzyme structure and mechanism to introduce specific mutations, enabling precise tailoring of enzyme properties for applications in drug development and synthetic biology. Combining directed evolution and rational design can yield synergistic improvements, enhancing enzyme efficiency and expanding their functional diversity in various biotechnological applications.
Advantages and Limitations of Directed Evolution
Directed evolution offers the advantage of generating proteins with enhanced functions through iterative rounds of mutation and selection without requiring detailed knowledge of protein structure or mechanisms. It is highly effective in exploring vast sequence space and identifying beneficial mutations that might be unpredictable by rational design. However, directed evolution can be time-consuming, resource-intensive, and may produce results that are difficult to interpret mechanistically, limiting its precision compared to rational design approaches.
Strengths and Challenges of Rational Design
Rational design leverages detailed structural and mechanistic knowledge of enzymes or proteins, enabling precise modifications to improve function or stability with fewer iterations compared to random methods. Its strengths lie in targeted specificity, predictability, and the ability to integrate computational modeling for designing variants with enhanced catalytic efficiency or altered substrate specificity. Challenges include the requirement for comprehensive understanding of structure-function relationships, limited success when structural data is incomplete, and potential difficulties in predicting complex protein dynamics or allosteric effects.
Integration of Computational Tools in Protein Engineering
Directed evolution leverages iterative rounds of mutation and selection to optimize protein functions, while rational design employs detailed structural and functional knowledge to engineer specific modifications. Integration of computational tools, such as molecular docking simulations, machine learning algorithms, and molecular dynamics, enhances both approaches by predicting mutation impacts and guiding library construction. These computational strategies accelerate protein engineering by reducing experimental trial-and-error and improving the accuracy of function predictions.
Case Studies: Success Stories and Lessons Learned
Directed evolution has enabled breakthroughs such as the development of more efficient enzymes for biofuel production by mimicking natural selection through iterative rounds of mutation and screening. Rational design has achieved success in creating novel protein drugs by leveraging computational modeling and structural biology to predict favorable mutations. Case studies reveal that combining directed evolution with rational design often yields synergistic improvements, highlighting the importance of integrating empirical data with theoretical insights for enhanced biomolecular engineering outcomes.
Future Perspectives and Emerging Trends
Directed evolution continues to accelerate enzyme optimization by mimicking natural selection for enhanced functionalities, while rational design leverages advances in computational modeling and AI-driven predictions to create tailored biomolecules with unprecedented precision. Integration of machine learning algorithms with high-throughput screening is emerging as a powerful trend, enabling more efficient exploration of vast protein sequence spaces. Future perspectives highlight hybrid approaches combining iterative mutagenesis with in silico design to overcome limitations of each method, driving innovation in synthetic biology, drug development, and industrial biotechnology.
Protein engineering
Directed evolution rapidly improves protein function by mimicking natural selection through iterative mutation and screening, while rational design uses detailed structural knowledge and computational models to precisely engineer proteins with desired properties.
Site-directed mutagenesis
Site-directed mutagenesis in directed evolution enables precise amino acid substitutions to rapidly generate protein variants, contrasting with rational design's reliance on detailed structural knowledge to predict beneficial mutations.
Combinatorial library
Directed evolution leverages combinatorial library generation to create vast genetic variants for enhanced protein function, whereas rational design relies on specific, knowledge-driven modifications without extensive library screening.
Enzyme optimization
Directed evolution rapidly enhances enzyme performance by mimicking natural selection through iterative mutation and screening, while rational design improves enzymes by applying structural and mechanistic knowledge to introduce targeted modifications for optimized catalytic efficiency.
Saturation mutagenesis
Saturation mutagenesis in directed evolution systematically explores all possible amino acid substitutions at specific protein sites, contrasting with rational design's targeted modifications based on structural predictions.
Molecular modeling
Molecular modeling enhances directed evolution by predicting mutational impacts on protein structure, while rational design leverages computational simulations to engineer novel enzymes with specific functions.
High-throughput screening
High-throughput screening accelerates directed evolution by rapidly analyzing vast enzyme variants, while rational design relies on limited mutant testing guided by structural insights.
Structure-guided design
Structure-guided design leverages detailed 3D protein structures to enable precise mutations in directed evolution, enhancing enzyme specificity and function more efficiently than trial-and-error approaches alone.
Functional selection
Functional selection in directed evolution rapidly identifies beneficial mutations from vast variant libraries, whereas rational design relies on structural knowledge to predict specific functional improvements.
Adaptive laboratory evolution
Adaptive laboratory evolution harnesses natural selection in controlled environments to enhance microbial traits more efficiently than rational design by exploring vast genetic diversity without prior mechanistic knowledge.
Directed evolution vs Rational design Infographic
 njnir.com
njnir.com