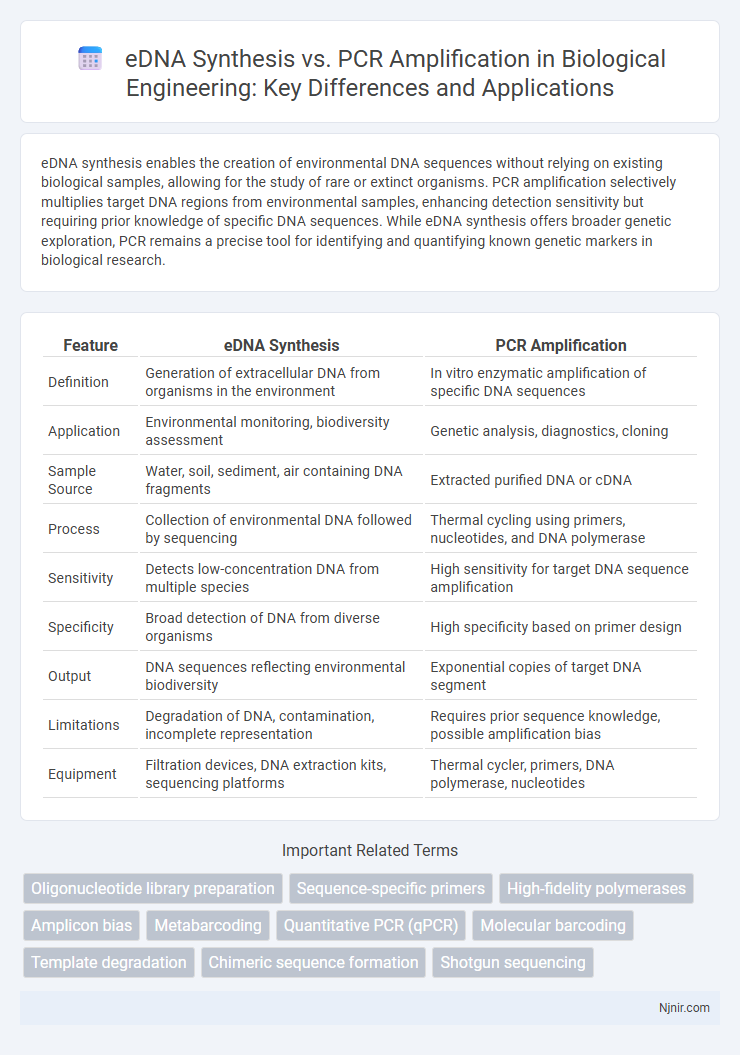
eDNA synthesis enables the creation of environmental DNA sequences without relying on existing biological samples, allowing for the study of rare or extinct organisms. PCR amplification selectively multiplies target DNA regions from environmental samples, enhancing detection sensitivity but requiring prior knowledge of specific DNA sequences. While eDNA synthesis offers broader genetic exploration, PCR remains a precise tool for identifying and quantifying known genetic markers in biological research.
Table of Comparison
| Feature | eDNA Synthesis | PCR Amplification |
|---|---|---|
| Definition | Generation of extracellular DNA from organisms in the environment | In vitro enzymatic amplification of specific DNA sequences |
| Application | Environmental monitoring, biodiversity assessment | Genetic analysis, diagnostics, cloning |
| Sample Source | Water, soil, sediment, air containing DNA fragments | Extracted purified DNA or cDNA |
| Process | Collection of environmental DNA followed by sequencing | Thermal cycling using primers, nucleotides, and DNA polymerase |
| Sensitivity | Detects low-concentration DNA from multiple species | High sensitivity for target DNA sequence amplification |
| Specificity | Broad detection of DNA from diverse organisms | High specificity based on primer design |
| Output | DNA sequences reflecting environmental biodiversity | Exponential copies of target DNA segment |
| Limitations | Degradation of DNA, contamination, incomplete representation | Requires prior sequence knowledge, possible amplification bias |
| Equipment | Filtration devices, DNA extraction kits, sequencing platforms | Thermal cycler, primers, DNA polymerase, nucleotides |
Overview of eDNA Synthesis and PCR Amplification
eDNA synthesis involves creating complementary DNA from environmental RNA samples using reverse transcriptase, enabling the study of gene expression in diverse microbial communities. PCR amplification selectively replicates specific DNA sequences through repetitive thermal cycling and enzyme activity, enhancing the detection and quantification of target genes. Both methods are essential for molecular ecology, with eDNA synthesis providing transcriptomic insights and PCR offering high sensitivity in DNA sequence analysis.
Fundamental Principles of eDNA Synthesis
eDNA synthesis involves creating complementary DNA strands from environmental RNA templates using reverse transcriptase, enabling the study of gene expression in environmental samples. This process requires specific primers and nucleotides to accurately transcribe RNA sequences into stable cDNA, preserving genetic information for downstream analysis. In contrast, PCR amplification targets existing DNA sequences, exponentially replicating specific DNA regions through thermal cycling and DNA polymerase activity.
PCR Amplification: Mechanisms and Applications
PCR amplification utilizes a cyclical process of denaturation, annealing, and extension to selectively replicate specific DNA sequences with high precision and efficiency. This method enables exponential amplification of target DNA fragments, making it indispensable for applications such as genetic fingerprinting, disease diagnostics, forensic analysis, and environmental DNA (eDNA) detection. The specificity and sensitivity of PCR amplification stem from primer design and thermal cycling, facilitating accurate quantification and identification of low-abundance genetic material in diverse biological samples.
Sensitivity and Specificity in eDNA and PCR Methods
Environmental DNA (eDNA) synthesis offers higher sensitivity than traditional PCR amplification due to its ability to detect low abundance genetic material in complex environmental samples, enabling early detection of rare or elusive species. PCR amplification, while sensitive, may suffer from nonspecific primer binding and amplification biases that reduce specificity and increase false positives, especially in heterogeneous eDNA mixtures. Optimizing primer design and using quantitative PCR (qPCR) or digital PCR platforms enhances specificity and quantification accuracy, making PCR methods robust but still generally less sensitive than comprehensive eDNA metabarcoding techniques.
Sample Collection and Preparation Considerations
Sample collection for eDNA synthesis requires meticulous avoidance of contamination and selection of appropriate substrates such as water, soil, or air filters to maximize DNA yield. Preparation involves immediate filtration or preservation using buffers or cold storage to prevent DNA degradation, whereas PCR amplification demands purified DNA extracts free of inhibitors like humic acids or phenolic compounds that can impede enzyme activity. Proper handling protocols and using suitable extraction kits tailored for environmental samples ensure reliable downstream PCR amplification and accurate biodiversity assessment.
Technical Challenges in eDNA Synthesis vs PCR
eDNA synthesis faces technical challenges such as low template concentration, DNA degradation, and environmental inhibitors that reduce synthesis efficiency and accuracy. PCR amplification struggles with primer specificity, amplification bias, and stochastic effects during low-copy-number DNA amplification, which can lead to false negatives or skewed abundance estimates. Both processes require meticulous optimization to overcome issues related to sensitivity, contamination, and reproducibility in environmental genetic analysis.
Comparative Efficiency for Biodiversity Monitoring
eDNA synthesis enables direct recovery of genetic material from environmental samples, offering higher sensitivity in detecting rare or elusive species compared to PCR amplification, which relies on target-specific primers and may exhibit amplification bias. PCR amplification is rapid and cost-effective for analyzing known species but can underestimate biodiversity due to primer mismatches and preferential amplification. Comparative studies show eDNA synthesis provides a more comprehensive representation of community diversity, enhancing accuracy and efficiency in biodiversity monitoring programs.
Cost Analysis and Resource Requirements
eDNA synthesis typically incurs higher upfront costs due to the need for specialized reagents and custom oligonucleotide design, whereas PCR amplification demands recurrent expenses primarily for primers, nucleotides, and thermal cycler operation. Resource requirements for eDNA synthesis include precision instrumentation and expert technical skills for accurate sequence assembly, contrasting with PCR's more accessible workflow and standard laboratory equipment. Cost-effectiveness of PCR amplification becomes evident in high-throughput or routine applications, while eDNA synthesis suits targeted applications requiring novel or synthetic genetic constructs despite increased resource intensity.
Future Trends in Molecular Detection Technologies
Future trends in molecular detection technologies emphasize the integration of environmental DNA (eDNA) synthesis with advanced PCR amplification techniques to enhance sensitivity and specificity in detecting low-abundance species. Innovations in digital PCR and next-generation sequencing (NGS) platforms are driving high-throughput eDNA analysis, enabling real-time monitoring of biodiversity and ecosystem health. Emerging microfluidic devices and machine learning algorithms further optimize assay workflows, reducing costs and processing times in environmental and clinical diagnostics.
Choosing the Right Approach for Biological Engineering
eDNA synthesis enables precise construction of genetic sequences for targeted modifications in biological engineering, offering high customization for synthetic biology applications. PCR amplification excels in rapidly replicating existing DNA segments, facilitating gene expression studies and mutation analysis with high sensitivity and efficiency. Selecting between eDNA synthesis and PCR amplification depends on project goals: de novo sequence creation requires eDNA synthesis, while amplifying known fragments favors PCR techniques.
Oligonucleotide library preparation
Oligonucleotide library preparation for eDNA synthesis requires precise sequence design and high-fidelity enzymatic assembly, whereas PCR amplification emphasizes primer specificity and efficient exponential DNA replication.
Sequence-specific primers
Sequence-specific primers in eDNA synthesis enable targeted amplification of genetic material, enhancing the sensitivity and specificity of PCR-based environmental DNA detection assays.
High-fidelity polymerases
High-fidelity polymerases enhance both eDNA synthesis and PCR amplification by minimizing replication errors, ensuring accurate DNA replication essential for precise genetic analysis.
Amplicon bias
Amplicon bias in eDNA synthesis and PCR amplification results in preferential amplification of certain DNA sequences, leading to skewed representation and reduced accuracy in biodiversity assessments.
Metabarcoding
Metabarcoding utilizes eDNA synthesis to capture diverse genetic material from environmental samples, while PCR amplification selectively targets and replicates specific DNA barcode regions to identify multiple species simultaneously.
Quantitative PCR (qPCR)
Quantitative PCR (qPCR) enables precise quantification of eDNA by amplifying target DNA sequences with fluorescent markers, offering higher sensitivity and specificity compared to traditional PCR amplification methods.
Molecular barcoding
Molecular barcoding enhances eDNA synthesis by uniquely tagging DNA fragments prior to PCR amplification, reducing amplification biases and enabling precise identification of species in environmental samples.
Template degradation
eDNA synthesis typically results in lower template degradation compared to PCR amplification, which subjects DNA to repeated thermal cycling that can cause significant template fragmentation and reduce target sequence integrity.
Chimeric sequence formation
eDNA synthesis often results in fewer chimeric sequences compared to PCR amplification, where the high number of cycles and template diversity significantly increase chimera formation.
Shotgun sequencing
Shotgun sequencing enables comprehensive genetic analysis by randomly shearing eDNA during synthesis, whereas PCR amplification selectively targets specific DNA regions, often limiting genomic coverage.
eDNA synthesis vs PCR amplification Infographic
 njnir.com
njnir.com