Cas9 and Cas12 are powerful gene-editing enzymes that differ in their DNA targeting mechanisms and cleavage patterns. Cas9 cleaves double-stranded DNA at specific sites using two distinct nuclease domains, while Cas12 introduces a staggered cut with a single nuclease domain and also exhibits nonspecific single-stranded DNase activity. These functional differences influence their applications in genome editing, diagnostics, and therapeutic development within biological engineering.
Table of Comparison
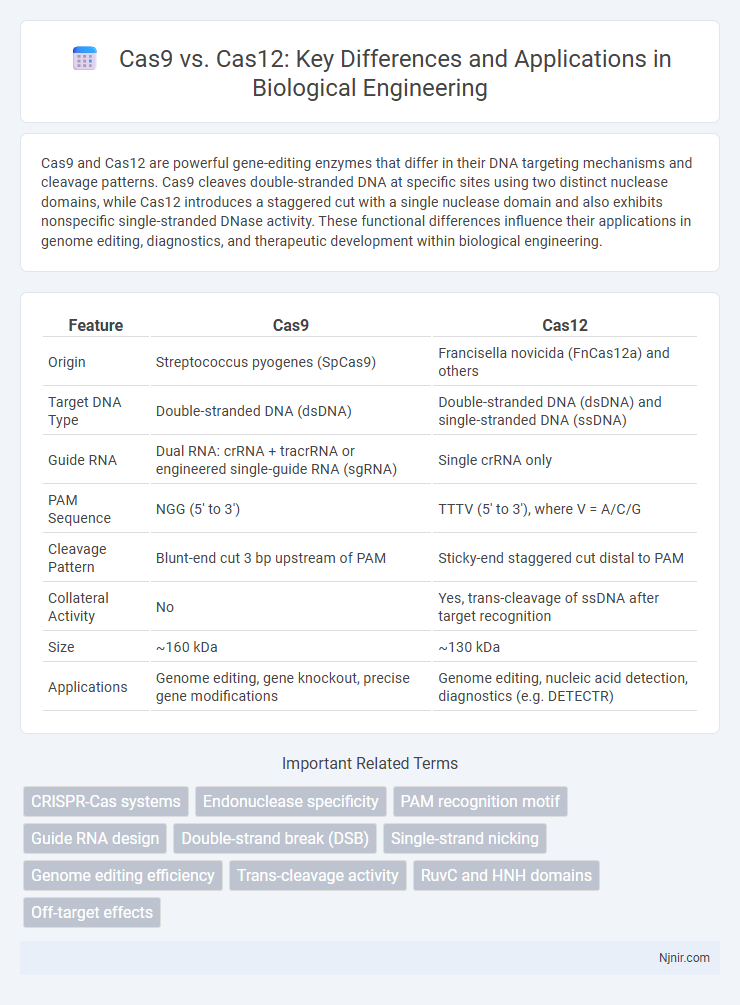
| Feature | Cas9 | Cas12 |
|---|---|---|
| Origin | Streptococcus pyogenes (SpCas9) | Francisella novicida (FnCas12a) and others |
| Target DNA Type | Double-stranded DNA (dsDNA) | Double-stranded DNA (dsDNA) and single-stranded DNA (ssDNA) |
| Guide RNA | Dual RNA: crRNA + tracrRNA or engineered single-guide RNA (sgRNA) | Single crRNA only |
| PAM Sequence | NGG (5' to 3') | TTTV (5' to 3'), where V = A/C/G |
| Cleavage Pattern | Blunt-end cut 3 bp upstream of PAM | Sticky-end staggered cut distal to PAM |
| Collateral Activity | No | Yes, trans-cleavage of ssDNA after target recognition |
| Size | ~160 kDa | ~130 kDa |
| Applications | Genome editing, gene knockout, precise gene modifications | Genome editing, nucleic acid detection, diagnostics (e.g. DETECTR) |
Overview of CRISPR-Cas Systems
The CRISPR-Cas systems utilize Cas9 and Cas12 as crucial DNA-targeting nucleases within adaptive immune defenses in bacteria and archaea. Cas9 recognizes and cleaves double-stranded DNA guided by a single-guide RNA, relying on a protospacer adjacent motif (PAM) sequence for target specificity. In contrast, Cas12 not only targets double-stranded DNA but also exhibits collateral single-stranded DNA cleavage upon activation, expanding its utility in genome editing and diagnostic applications.
Structural Differences Between Cas9 and Cas12
Cas9 and Cas12 exhibit distinct structural differences influencing their DNA targeting and cleavage mechanisms. Cas9 possesses two nuclease domains, RuvC and HNH, that cleave complementary and non-complementary DNA strands respectively, while Cas12 contains a single RuvC domain responsible for cleaving both DNA strands. Cas12 also features a prominent PAM-interacting domain and a unique single RNA guide structure, differentiating it from the dual-guide RNA requirement seen in Cas9 systems.
Mechanisms of DNA Recognition and Cleavage
Cas9 recognizes DNA by a guide RNA that pairs with a complementary target sequence adjacent to a protospacer adjacent motif (PAM), inducing a conformational change that activates two nuclease domains, RuvC and HNH, to create a double-stranded break. Cas12 also utilizes a guide RNA for target recognition but binds a T-rich PAM and induces a conformational change that activates a single RuvC-like domain responsible for cleaving both DNA strands and exhibits collateral single-stranded DNA cleavage activity. The structural differences in PAM recognition and nuclease domain activation between Cas9 and Cas12 result in distinct DNA cleavage patterns and specificity profiles.
Guide RNA Requirements: Cas9 vs Cas12
Cas9 requires a single guide RNA (sgRNA) that combines CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) sequences for DNA targeting, typically recognizing a 20-nucleotide complementary sequence adjacent to a PAM site (5'-NGG-3'). Cas12 uses a shorter, simpler crRNA without the need for tracrRNA and targets a different PAM sequence, often 5'-TTTV-3', enabling cleavage of double-stranded DNA with a distinct mechanism. These guide RNA requirements affect specificity, target range, and application flexibility in genome editing technologies.
Specificity and Off-Target Effects
Cas9 exhibits high efficiency in gene editing but can show off-target effects due to its less stringent PAM sequence requirements, leading to potential unintended DNA cleavage. Cas12 offers improved specificity through a longer PAM sequence and a distinct DNA recognition mechanism, reducing off-target activity and enhancing precision in genome editing. Advances in engineered Cas9 variants and Cas12 orthologs continue to minimize off-target risks while maintaining robust target specificity.
Applications in Genome Editing
Cas9 and Cas12 are prominent CRISPR-associated nucleases used for precise genome editing in various organisms, with Cas9 predominantly targeting double-stranded DNA and Cas12 capable of both DNA and single-stranded DNA cleavage. Cas9's widespread use in gene knockout, gene correction, and transcriptional regulation is attributed to its well-characterized PAM sequence and robust cleavage activity, while Cas12 offers advantages in detecting genetic variations and enabling multiplexed editing due to its collateral cleavage activity. Applications in therapeutic development, functional genomics, and agricultural biotechnology exploit Cas9's precision and Cas12's versatility to advance genome modification strategies.
Efficiency of Gene Knockout and Knock-In
Cas9 exhibits high efficiency in gene knockout due to its precise double-strand breaks and well-established PAM requirements, facilitating targeted gene disruption. Cas12 offers enhanced versatility in knock-in applications through its ability to generate staggered cuts, promoting more efficient homology-directed repair and improving insertion precision. Comparative studies reveal Cas12's lower off-target activity, which contributes to increased gene editing accuracy in both knockout and knock-in workflows.
Multiplexing Capabilities: Comparative Analysis
Cas9 and Cas12 exhibit distinct multiplexing capabilities crucial for genetic engineering applications. Cas9 can simultaneously target multiple DNA sequences through the use of multiple guide RNAs (gRNAs), but its reliance on a PAM sequence restricts target site selection; Cas12 offers greater flexibility with a T-rich PAM and cleavage of single-stranded DNA, enhancing multiplex editing efficiency. Comparative studies demonstrate Cas12's superior performance in complex genome editing scenarios, enabling higher throughput and precision in multiplexed gene modifications.
Delivery Methods in Biological Systems
Cas9 and Cas12 nucleases differ significantly in their delivery methods in biological systems, with Cas9 frequently delivered via adeno-associated viruses (AAV) and lipid nanoparticles due to its well-characterized size and stability. Cas12, often smaller and featuring distinct cleavage properties, enables more versatile delivery through both viral vectors like lentivirus and non-viral systems including electroporation and synthetic nanoparticles. The choice between Cas9 and Cas12 delivery depends on target cell type, genome-editing efficiency, and potential off-target effects, making vector size and cellular uptake critical factors in optimizing therapeutic applications.
Future Prospects in Synthetic Biology
Cas9 and Cas12 enzymes offer distinct advantages for future synthetic biology applications, with Cas9 excelling in precise genome editing due to its well-characterized PAM recognition and high specificity. Cas12 shows promise for multiplexed editing and diagnostics through its ability to process CRISPR arrays and collateral cleavage activity, enabling advanced synthetic circuits and biosensors. Ongoing engineering of both nucleases aims to expand targeting ranges, reduce off-target effects, and improve delivery methods for therapeutic gene editing and programmable cellular functions.
CRISPR-Cas systems
Cas9 and Cas12 are CRISPR-Cas systems characterized by distinct PAM recognition sequences, differing DNA cleavage mechanisms, and varying applications in genome editing technologies.
Endonuclease specificity
Cas9 endonucleases exhibit high specificity by targeting a 20-nucleotide DNA sequence adjacent to a PAM site, while Cas12 endonucleases recognize a different PAM and cleave with broader target flexibility but increased off-target potential.
PAM recognition motif
Cas9 enzymes recognize a PAM sequence typically characterized by the NGG motif, while Cas12 enzymes target a more diverse PAM sequence, often T-rich such as TTTV, enabling distinct DNA targeting specificities.
Guide RNA design
Cas12 guide RNA design requires a longer CRISPR RNA with a distinct T-rich PAM sequence compared to Cas9, which utilizes a shorter single-guide RNA targeting an NGG PAM for precise DNA cleavage.
Double-strand break (DSB)
Cas9 creates blunt-ended double-strand breaks (DSBs) typically 3 base pairs upstream of the PAM site, whereas Cas12 induces staggered DSBs with sticky overhangs distal to the PAM, influencing DNA repair outcomes.
Single-strand nicking
Cas12 exhibits robust single-strand nicking activity on target DNA strands, whereas Cas9 primarily induces double-strand breaks with limited capacity for precise single-strand nicking.
Genome editing efficiency
Cas9 generally exhibits higher genome editing efficiency compared to Cas12 due to its precise DNA cleavage and strong target specificity.
Trans-cleavage activity
Cas12 exhibits robust trans-cleavage activity by indiscriminately cutting single-stranded DNA upon target recognition, whereas Cas9 lacks this trans-cleavage capability, making Cas12 more effective for biosensing applications.
RuvC and HNH domains
Cas9 contains both RuvC and HNH nuclease domains enabling it to create double-strand breaks at specific DNA sites, while Cas12 possesses only the RuvC domain, which cleaves DNA via a distinct single-strand cleavage mechanism.
Off-target effects
Cas12 exhibits lower off-target effects than Cas9 due to its higher sequence specificity and unique PAM recognition.
Cas9 vs Cas12 Infographic
 njnir.com
njnir.com