Microarray analysis enables the simultaneous examination of gene expression by hybridizing labeled cDNA to predefined probes on a chip, offering cost-effective and rapid profiling of known transcripts. Next-generation sequencing (NGS) provides a comprehensive and unbiased approach by directly sequencing millions of DNA fragments, allowing detection of novel transcripts, mutations, and splice variants with higher sensitivity and resolution. The choice between these technologies depends on the specificity of research goals, budget constraints, and the need for discovery versus targeted analysis in biomedical engineering applications.
Table of Comparison
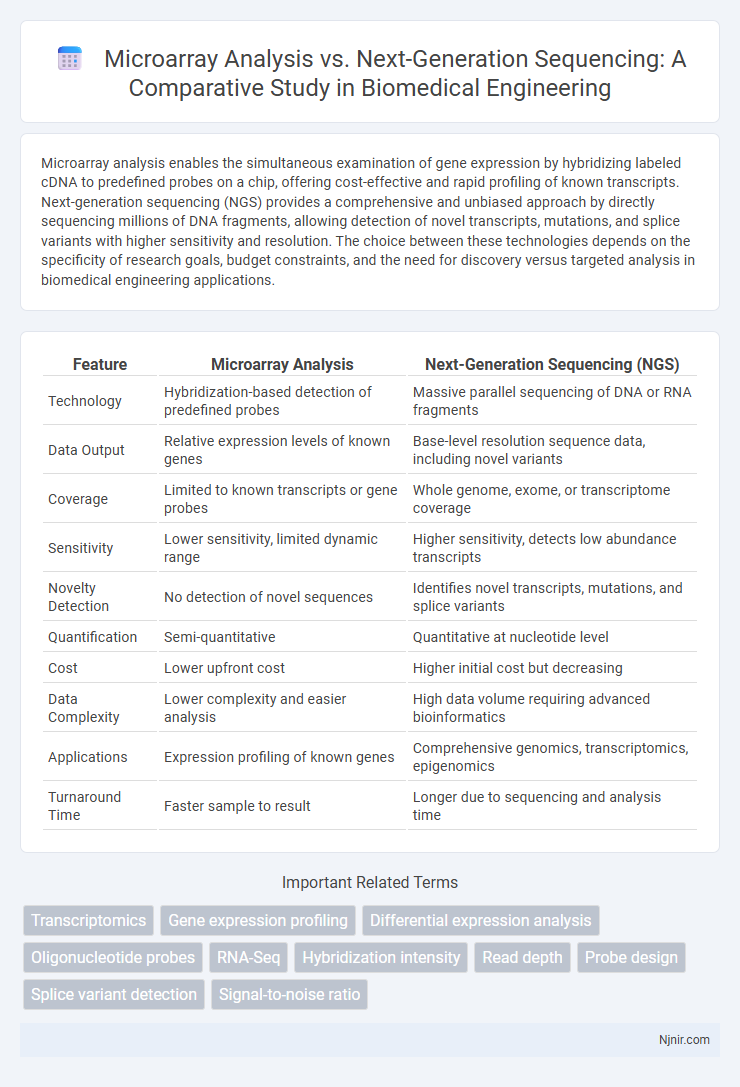
| Feature | Microarray Analysis | Next-Generation Sequencing (NGS) |
|---|---|---|
| Technology | Hybridization-based detection of predefined probes | Massive parallel sequencing of DNA or RNA fragments |
| Data Output | Relative expression levels of known genes | Base-level resolution sequence data, including novel variants |
| Coverage | Limited to known transcripts or gene probes | Whole genome, exome, or transcriptome coverage |
| Sensitivity | Lower sensitivity, limited dynamic range | Higher sensitivity, detects low abundance transcripts |
| Novelty Detection | No detection of novel sequences | Identifies novel transcripts, mutations, and splice variants |
| Quantification | Semi-quantitative | Quantitative at nucleotide level |
| Cost | Lower upfront cost | Higher initial cost but decreasing |
| Data Complexity | Lower complexity and easier analysis | High data volume requiring advanced bioinformatics |
| Applications | Expression profiling of known genes | Comprehensive genomics, transcriptomics, epigenomics |
| Turnaround Time | Faster sample to result | Longer due to sequencing and analysis time |
Introduction to Microarray Analysis and Next-Generation Sequencing
Microarray analysis enables simultaneous measurement of gene expression levels across thousands of genes by hybridizing labeled cDNA to pre-designed probes on a solid surface. Next-generation sequencing (NGS) offers high-throughput sequencing of entire genomes or transcriptomes, providing detailed information about mutations, gene expression, and epigenetic modifications. Both technologies are pivotal in genomics research, with microarrays focusing on known sequences and NGS delivering broader, unbiased data sets for comprehensive analysis.
Fundamental Principles: Microarray vs NGS Technologies
Microarray analysis relies on hybridization of fluorescently labeled DNA or RNA samples to predefined probes on a solid surface, enabling detection of known sequences through intensity signals. Next-generation sequencing (NGS) technology sequences millions of DNA fragments in parallel by synthesizing or detecting nucleotide incorporation, providing comprehensive data on sequence variation and expression profiles. While microarrays are limited by probe design and detect only targeted sequences, NGS offers unbiased, high-resolution analysis of both known and novel genetic information.
Workflow Comparison: Sample Preparation and Data Acquisition
Microarray analysis involves hybridizing fluorescently labeled DNA or RNA samples to predefined probes on a chip, requiring extensive sample amplification and labeling steps before scanning for fluorescence intensity data. Next-generation sequencing (NGS) workflows begin with fragmenting DNA or RNA, followed by library preparation including adapter ligation, clonal amplification, and high-throughput sequencing to generate raw sequence reads. Data acquisition in microarrays is limited to known probes and relative expression levels, whereas NGS provides comprehensive, base-level resolution enabling detection of novel sequences and quantitative analysis of entire transcriptomes or genomes.
Sensitivity and Specificity: Analytical Performance
Next-generation sequencing offers higher sensitivity compared to microarray analysis, enabling the detection of low-abundance transcripts and rare genetic variants with greater accuracy. Microarrays typically have limited dynamic range and cross-hybridization can reduce specificity, whereas NGS provides base-level resolution and minimizes false positives through precise alignment algorithms. Analytical performance studies consistently show NGS achieving superior specificity and sensitivity metrics, making it a preferred choice for comprehensive genomic profiling.
Applications in Biomedical Engineering and Research
Microarray analysis enables high-throughput examination of gene expression profiles and genetic variations, making it valuable for biomarker discovery and disease diagnostics in biomedical engineering. Next-generation sequencing (NGS) offers comprehensive genomic and transcriptomic insights with higher resolution, facilitating applications such as personalized medicine, cancer genomics, and complex disease modeling. Both technologies drive advancements in systems biology, drug development, and regenerative medicine by providing critical data for understanding molecular mechanisms and therapeutic targets.
Data Output and Bioinformatics Challenges
Microarray analysis generates fixed, predefined probe data reflecting gene expression levels, allowing for limited detection of known transcripts but producing smaller, more structured datasets compared to next-generation sequencing (NGS), which provides comprehensive, high-throughput reads enabling detection of novel variants, isoforms, and low-abundance transcripts. NGS data output is exponentially larger and more complex, requiring advanced bioinformatics tools for alignment, assembly, variant calling, and annotation, whereas microarray data demands robust normalization and background correction methods but involves fewer computational resources. Bioinformatics challenges in NGS include managing vast data storage, ensuring accurate short-read mapping, and addressing sequencing errors, while microarray analysis faces obstacles in probe specificity and cross-hybridization effects limiting data precision.
Cost-Effectiveness and Scalability
Microarray analysis offers a lower upfront cost and simpler workflow, making it cost-effective for targeted gene expression studies across moderate sample sizes. Next-generation sequencing (NGS) provides superior scalability and data depth, efficiently processing large sample cohorts with comprehensive genomic information despite higher initial expenses. Laboratories prioritizing extensive genome-wide analysis and future scalability often find NGS a more economically viable choice over time.
Recent Advances and Hybrid Approaches
Microarray analysis offers high-throughput gene expression profiling, but next-generation sequencing (NGS) provides deeper insights with greater sensitivity and the ability to detect novel transcripts and mutations. Recent advances in NGS technology include improved accuracy, reduced costs, and enhanced bioinformatics tools, enabling more comprehensive genomic and transcriptomic analyses. Hybrid approaches combining microarray and NGS leverage the cost-effectiveness of microarrays with the detailed resolution of sequencing, optimizing data interpretation in complex biological studies.
Limitations and Potential Pitfalls
Microarray analysis faces limitations such as lower sensitivity, restricted to known sequences, and cross-hybridization errors, which can lead to inaccurate gene expression quantification. Next-generation sequencing (NGS) overcomes many microarray constraints by offering higher resolution and detecting novel transcripts but encounters challenges like complex data analysis, sequencing biases, and higher costs. Both technologies require careful experimental design and validation strategies to mitigate technical artifacts and ensure reliable genomic insights.
Future Perspectives in Biomarker Discovery and Precision Medicine
Next-generation sequencing (NGS) offers higher resolution and greater sensitivity compared to microarray analysis, enabling comprehensive detection of rare genetic variants and novel biomarkers crucial for precision medicine. Advances in NGS technologies facilitate integrative multi-omics approaches, fostering personalized diagnostics and targeted therapies by uncovering complex molecular profiles. Emerging trends highlight the potential for NGS to surpass microarrays in scalability and cost-effectiveness, accelerating biomarker discovery pipelines and enhancing clinical decision-making in future healthcare frameworks.
Transcriptomics
Next-generation sequencing offers higher sensitivity, broader dynamic range, and more comprehensive transcriptomic profiling compared to microarray analysis, enabling detection of novel transcripts, alternative splicing, and low-abundance RNAs.
Gene expression profiling
Next-generation sequencing provides more comprehensive and quantitative gene expression profiling than microarray analysis by detecting a wider range of transcripts, including rare and novel variants, with higher sensitivity and dynamic range.
Differential expression analysis
Next-generation sequencing provides higher sensitivity and broader dynamic range than microarray analysis for differential expression analysis, enabling more accurate detection of low-abundance transcripts and novel gene variants.
Oligonucleotide probes
Microarray analysis relies on fixed oligonucleotide probes for hybridization-based detection of known sequences, whereas next-generation sequencing uses random priming and sequencing by synthesis to identify both known and novel sequences without predefined probes.
RNA-Seq
RNA-Seq in next-generation sequencing provides higher sensitivity, greater dynamic range, and more precise quantification of transcriptomes compared to traditional microarray analysis.
Hybridization intensity
Microarray analysis quantifies gene expression through hybridization intensity signals on predefined probes, while Next-generation sequencing directly sequences cDNA, providing base-level resolution without relying on hybridization intensity measurements.
Read depth
Next-generation sequencing offers significantly higher read depth than microarray analysis, enabling more precise detection of rare variants and low-abundance transcripts.
Probe design
Next-generation sequencing eliminates reliance on predefined probe design required in microarray analysis, enabling unbiased, high-resolution detection of genetic variations and expression levels.
Splice variant detection
Next-generation sequencing provides higher resolution and accuracy than microarray analysis for detecting splice variants by directly sequencing RNA transcripts and identifying novel or low-abundance isoforms.
Signal-to-noise ratio
Next-generation sequencing offers a higher signal-to-noise ratio than microarray analysis by providing more precise and quantitative detection of low-abundance transcripts through direct nucleotide sequencing.
Microarray analysis vs Next-generation sequencing Infographic
 njnir.com
njnir.com