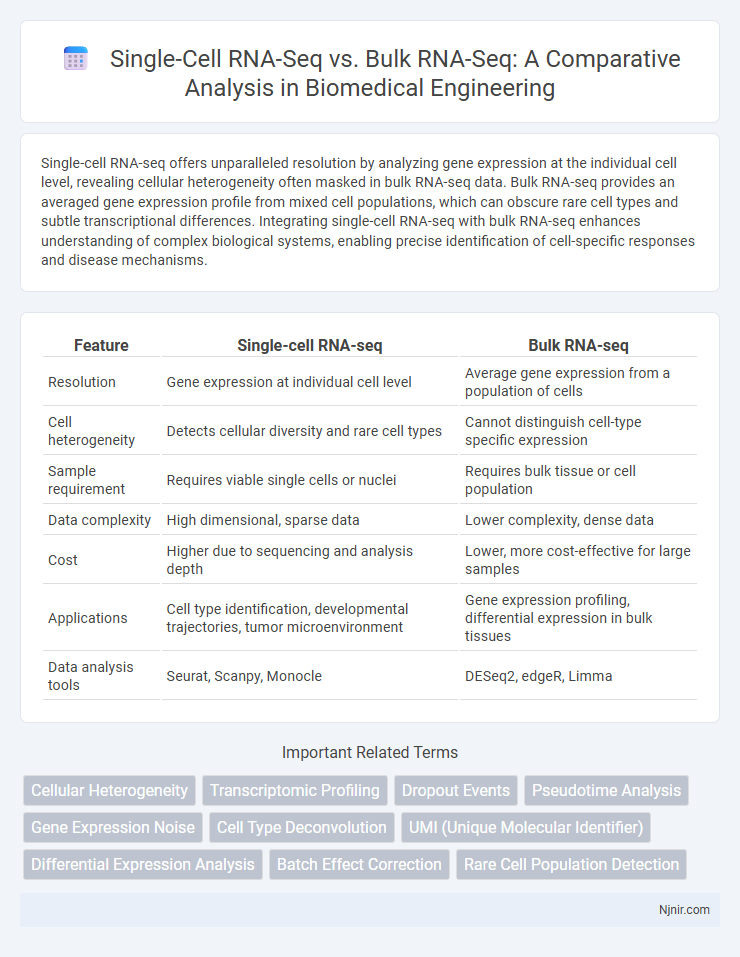
Single-cell RNA-seq offers unparalleled resolution by analyzing gene expression at the individual cell level, revealing cellular heterogeneity often masked in bulk RNA-seq data. Bulk RNA-seq provides an averaged gene expression profile from mixed cell populations, which can obscure rare cell types and subtle transcriptional differences. Integrating single-cell RNA-seq with bulk RNA-seq enhances understanding of complex biological systems, enabling precise identification of cell-specific responses and disease mechanisms.
Table of Comparison
| Feature | Single-cell RNA-seq | Bulk RNA-seq |
|---|---|---|
| Resolution | Gene expression at individual cell level | Average gene expression from a population of cells |
| Cell heterogeneity | Detects cellular diversity and rare cell types | Cannot distinguish cell-type specific expression |
| Sample requirement | Requires viable single cells or nuclei | Requires bulk tissue or cell population |
| Data complexity | High dimensional, sparse data | Lower complexity, dense data |
| Cost | Higher due to sequencing and analysis depth | Lower, more cost-effective for large samples |
| Applications | Cell type identification, developmental trajectories, tumor microenvironment | Gene expression profiling, differential expression in bulk tissues |
| Data analysis tools | Seurat, Scanpy, Monocle | DESeq2, edgeR, Limma |
Introduction to RNA Sequencing Technologies
Single-cell RNA sequencing (scRNA-seq) enables gene expression analysis at the individual cell level, revealing cellular heterogeneity and rare cell populations. Bulk RNA sequencing (Bulk RNA-seq) measures average gene expression across a mixed population of cells, providing overall transcriptomic insights. Both technologies utilize next-generation sequencing platforms but differ in resolution, sample preparation, and data complexity.
Principles of Bulk RNA-seq
Bulk RNA-seq measures the aggregate gene expression of thousands to millions of cells, providing an averaged transcriptomic profile that reflects dominant cellular populations. The methodology involves extracting RNA from homogenized tissue samples, converting it into cDNA, and sequencing to quantify transcript abundance across the sample. This approach is cost-effective and suitable for detecting overall expression trends but lacks cellular resolution and masks heterogeneity present in complex tissues.
Fundamentals of Single-cell RNA-seq
Single-cell RNA sequencing (scRNA-seq) enables the analysis of gene expression at the individual cell level, revealing cellular heterogeneity that bulk RNA-seq cannot capture. By isolating and sequencing RNA from thousands of single cells, scRNA-seq provides insights into cell type diversity, developmental trajectories, and cellular responses within complex tissues. This technique relies on microfluidics or droplet-based technologies to partition cells, followed by reverse transcription and amplification of cDNA, enabling high-resolution transcriptomic profiling crucial for understanding cellular function and disease mechanisms.
Key Differences Between Single-cell and Bulk RNA-seq
Single-cell RNA-seq (scRNA-seq) profiles gene expression at the individual cell level, capturing cellular heterogeneity and rare cell populations often masked in bulk RNA-seq. Bulk RNA-seq provides averaged gene expression data from mixed cell populations, losing resolution on cell-specific transcriptional differences. The key differences include sensitivity to cellular diversity, resolution of cell types, and data complexity, with scRNA-seq enabling insights into cellular dynamics and developmental processes that bulk RNA-seq cannot resolve.
Applications in Biomedical Research
Single-cell RNA-seq enables precise analysis of gene expression at an individual cell level, revealing cellular heterogeneity and rare cell populations crucial for understanding complex tissues and disease mechanisms. Bulk RNA-seq provides averaged gene expression profiles from mixed cell populations, useful for identifying overall transcriptional changes in samples such as tumors or treated tissues. Applications in biomedical research include tumor microenvironment studies, immune cell profiling in disease, and drug response analysis, where single-cell RNA-seq offers high-resolution insights while bulk RNA-seq supports population-level gene expression trends.
Data Analysis and Interpretation Challenges
Single-cell RNA-seq data analysis faces challenges including high dimensionality, dropout events, and biological variability requiring specialized normalization and imputation techniques. Bulk RNA-seq averages gene expression across heterogeneous cell populations, simplifying analysis but obscuring cell-type-specific signals and limiting resolution. Interpretation of single-cell data demands advanced computational tools like clustering algorithms and trajectory inference to unravel cellular heterogeneity, while bulk RNA-seq relies on robust statistical models for differential expression analysis at the population level.
Advantages of Single-cell RNA-seq
Single-cell RNA-seq enables the analysis of gene expression at an individual cell level, revealing cellular heterogeneity and rare cell populations undetectable by bulk RNA-seq. This technique allows for precise identification of distinct cell types, states, and developmental trajectories within complex tissues. Single-cell RNA-seq provides higher resolution data that enhances understanding of cellular diversity and dynamic biological processes.
Limitations of Bulk RNA-seq
Bulk RNA-seq averages gene expression across thousands of cells, masking cellular heterogeneity and rare cell populations, which limits insights into cell-type-specific transcriptional profiles. It cannot resolve dynamic cellular states or identify gene expression variability at the single-cell level, reducing sensitivity to subtle but biologically significant differences. These limitations hinder the understanding of complex tissues, developmental processes, and disease mechanisms that rely on cell-to-cell variability.
Emerging Trends and Innovations
Single-cell RNA-seq enables high-resolution gene expression profiling by capturing cellular heterogeneity, whereas Bulk RNA-seq averages signals across cell populations, potentially masking rare cell types. Emerging innovations in single-cell RNA-seq include multi-omics integration, spatial transcriptomics, and improved protocols for higher throughput and sensitivity. Advances in computational methods, such as machine learning algorithms for data deconvolution, are enhancing the interpretation of Bulk RNA-seq datasets, bridging the gap between single-cell resolution and bulk sample analysis.
Future Directions in Biomedical Engineering
Single-cell RNA-seq enables precise characterization of cellular heterogeneity, offering opportunities for personalized diagnostics and targeted therapies in biomedical engineering. Future advancements will focus on integrating multi-omics data and improving computational methods for high-resolution spatial transcriptomics. Bulk RNA-seq remains valuable for population-level gene expression profiling, complementing single-cell approaches to provide comprehensive insights into tissue-level dynamics.
Cellular Heterogeneity
Single-cell RNA-seq enables precise characterization of cellular heterogeneity by profiling gene expression at the individual cell level, whereas Bulk RNA-seq averages signals across mixed cell populations, masking distinct cellular contributions.
Transcriptomic Profiling
Single-cell RNA-seq provides high-resolution transcriptomic profiling by capturing gene expression at the individual cell level, whereas bulk RNA-seq measures average gene expression across heterogeneous cell populations, potentially masking cellular heterogeneity.
Dropout Events
Single-cell RNA-seq exhibits higher dropout events than bulk RNA-seq due to low mRNA capture efficiency and stochastic gene expression, impacting transcript detection sensitivity.
Pseudotime Analysis
Single-cell RNA-seq enables high-resolution pseudotime analysis by capturing transcriptomic heterogeneity at the individual cell level, unlike bulk RNA-seq which averages gene expression across cell populations and obscures dynamic cellular trajectories.
Gene Expression Noise
Single-cell RNA-seq captures gene expression noise at the individual cell level, revealing cellular heterogeneity, whereas bulk RNA-seq averages signals across populations, masking this variability.
Cell Type Deconvolution
Single-cell RNA-seq provides precise cell type deconvolution by profiling individual cell transcriptomes, whereas Bulk RNA-seq captures averaged gene expression across mixed cell populations, requiring computational methods to infer cell type proportions.
UMI (Unique Molecular Identifier)
Single-cell RNA-seq employs UMIs to accurately quantify individual transcript copies, reducing amplification bias, while bulk RNA-seq typically lacks UMI integration, limiting precise molecule counting.
Differential Expression Analysis
Single-cell RNA-seq enables identification of cell-specific differential gene expression patterns, whereas bulk RNA-seq provides averaged expression profiles across heterogeneous cell populations, limiting resolution in differential expression analysis.
Batch Effect Correction
Single-cell RNA-seq batch effect correction requires advanced algorithms like Harmony or Seurat's integration method to address high cellular heterogeneity, whereas bulk RNA-seq often uses ComBat or limma for more homogeneous sample adjustments.
Rare Cell Population Detection
Single-cell RNA-seq enables precise detection of rare cell populations by analyzing gene expression at the individual cell level, unlike Bulk RNA-seq which averages signals across heterogeneous samples, often masking low-abundance cell types.
Single-cell RNA-seq vs Bulk RNA-seq Infographic
 njnir.com
njnir.com